
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,543,487 – 7,543,664 |
| Length | 177 |
| Max. P | 0.999134 |

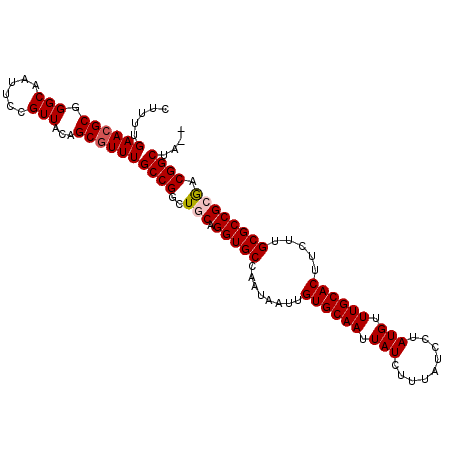
| Location | 7,543,487 – 7,543,592 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 95.27 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -29.14 |
| Energy contribution | -29.58 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7543487 105 + 20766785 CUUUUGAACGCGGGCAAUUCCGUUACAGCGUUUGCCGGCUCCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCCUAUGUUUGCACUUCUUGCGCCGCGACGGCUA-- .....((((((.(((......)))...))))))((((......(((((........((((((.................)))))).....)))))....))))..-- ( -29.25) >DroSec_CAF1 1733 105 + 1 UUUUUGAACGCGGGCAAUUCCGUUACAGCGUUUGCCGGCUGCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCCUAUGUUUGCACUUCUUGCGCCGAGACGGCUA-- .....((((((.(((......)))...))))))((((.((...(((((........((((((.................)))))).....))))).)).))))..-- ( -29.95) >DroSim_CAF1 1741 105 + 1 UUUUUGAACGCGGGCAAUUCCGUUACAGCGUUUGCCGGCUGCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCCUAUGUUUGCACUUCUUGCGCCGCGACGGCUA-- .....((((((.(((......)))...))))))((((..(((.(((((........((((((.................)))))).....)))))))).))))..-- ( -32.55) >DroEre_CAF1 4627 107 + 1 CUUUUGAACGCGGGCAAUUCCGUUACGGCUUUUGCCGGCUGCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCUUAUGUUUGCACUUCUUGCGCCGCGACGGCAAUU .........(((((....)))))..((((....))))(((((.(((((........((((((.................)))))).....)))))..).)))).... ( -30.85) >DroYak_CAF1 1738 105 + 1 CUUUUGAACGCGGGCAAUUCCGUUACGGCGUUUGCCGGCUGCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCCUAUGCUUGCACUUCUUGCGCCGCAACGGCAA-- .....((((((.(............).))))))((((..(((.(((((........((((((.(((.........))).)))))).....)))))))).))))..-- ( -32.92) >consensus CUUUUGAACGCGGGCAAUUCCGUUACAGCGUUUGCCGGCUGCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCCUAUGUUUGCACUUCUUGCGCCGCGACGGCUA__ .....((((((.(((......)))...))))))((((..(((.(((((........((((((.(((.........))).)))))).....)))))))).)))).... (-29.14 = -29.58 + 0.44)

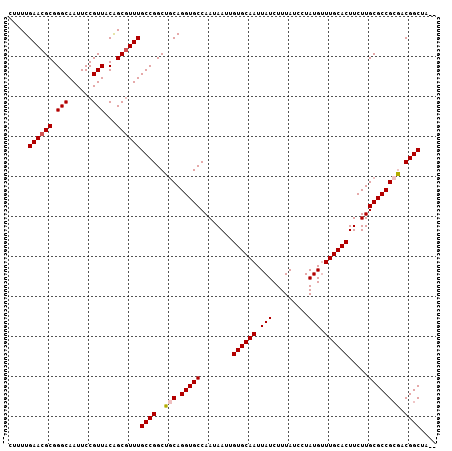
| Location | 7,543,487 – 7,543,592 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 95.27 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -25.81 |
| Energy contribution | -25.77 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7543487 105 - 20766785 --UAGCCGUCGCGGCGCAAGAAGUGCAAACAUAGGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGGAGCCGGCAAACGCUGUAACGGAAUUGCCCGCGUUCAAAAG --.....(.(((((.((((...((((..............(((((((....))))))).))))(((..((.(((....)))))..)))..)))))))))..)..... ( -26.96) >DroSec_CAF1 1733 105 - 1 --UAGCCGUCUCGGCGCAAGAAGUGCAAACAUAGGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGCAGCCGGCAAACGCUGUAACGGAAUUGCCCGCGUUCAAAAA --..((((...))))((.....((((((.................)))))).......((((((((((((.(.....)))))))..))...)))).))......... ( -26.33) >DroSim_CAF1 1741 105 - 1 --UAGCCGUCGCGGCGCAAGAAGUGCAAACAUAGGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGCAGCCGGCAAACGCUGUAACGGAAUUGCCCGCGUUCAAAAA --.....(.(((((.((((...((((((.................))))))...........((((((((.(.....)))))))..))..)))))))))..)..... ( -28.13) >DroEre_CAF1 4627 107 - 1 AAUUGCCGUCGCGGCGCAAGAAGUGCAAACAUAAGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGCAGCCGGCAAAAGCCGUAACGGAAUUGCCCGCGUUCAAAAG ..(((....(((((.((((...((((((.................)))))).......(((.......)))(((....))).........)))))))))..)))... ( -28.73) >DroYak_CAF1 1738 105 - 1 --UUGCCGUUGCGGCGCAAGAAGUGCAAGCAUAGGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGCAGCCGGCAAACGCCGUAACGGAAUUGCCCGCGUUCAAAAG --...(((((((((((......((((((.................)))))).....(((((.......)))))....)))))))))))................... ( -32.53) >consensus __UAGCCGUCGCGGCGCAAGAAGUGCAAACAUAGGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGCAGCCGGCAAACGCUGUAACGGAAUUGCCCGCGUUCAAAAG .....((((..(((((......((((((.................)))))).....(((((.......)))))....)))))..))))................... (-25.81 = -25.77 + -0.04)

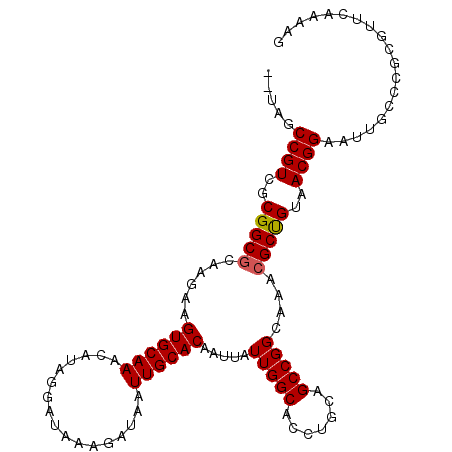

| Location | 7,543,516 – 7,543,627 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 94.55 |
| Mean single sequence MFE | -38.18 |
| Consensus MFE | -33.12 |
| Energy contribution | -33.00 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7543516 111 + 20766785 GUUUGCCGGCUCCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCCUAUGUUUGCACUUCUUGCGCCGCGACGGCUA-------UGUGGCAACGCUACAGCUGUUUGCCAAGCACAACA ((((((.(((....(((((........((((((.................)))))).....)))))..(((((((.-------.(((....)))...))))))).)))..))).))). ( -35.35) >DroSec_CAF1 1762 111 + 1 GUUUGCCGGCUGCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCCUAUGUUUGCACUUCUUGCGCCGAGACGGCUA-------UGUGGCAACGCCACAGCUGUUUGCCAAGCACAACA ((((((.(((....(((((........((((((.................)))))).....))))).((((((((.-------.(((((...))))))))))))))))..))).))). ( -37.75) >DroSim_CAF1 1770 111 + 1 GUUUGCCGGCUGCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCCUAUGUUUGCACUUCUUGCGCCGCGACGGCUA-------UGUGGCAACGCCACAGCUGUUUGCCAAGCACAACA ((((((.(((.((.(((((........((((((.................)))))).....)))))))(((((((.-------.(((((...)))))))))))).)))..))).))). ( -38.25) >DroEre_CAF1 4656 118 + 1 UUUUGCCGGCUGCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCUUAUGUUUGCACUUCUUGCGCCGCGACGGCAAUUGGCAGUGUGGCAACGCCACAGCUGUUUGCCAAGCACAACA ..((((((..(((.(((((........((((((.................)))))).....)))))))).))))))(((((((((((((...)))))).....)))))))........ ( -41.55) >DroYak_CAF1 1767 111 + 1 GUUUGCCGGCUGCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCCUAUGCUUGCACUUCUUGCGCCGCAACGGCAA-------UGUGGCAACGCCACAGCUGUUUGCCAAGCACAACA ..((((((..(((.(((((........((((((.(((.........))).)))))).....)))))))).))))))-------((((((...))))))(((........)))...... ( -38.02) >consensus GUUUGCCGGCUGCAGGUGCCAAUAAUUGUGCAAUUAUCUUUAUCCUAUGUUUGCACUUCUUGCGCCGCGACGGCUA_______UGUGGCAACGCCACAGCUGUUUGCCAAGCACAACA ((((((.(((....(((((........((((((.(((.........))).)))))).....)))))..((((((.........((((((...)))))))))))).)))..))).))). (-33.12 = -33.00 + -0.12)

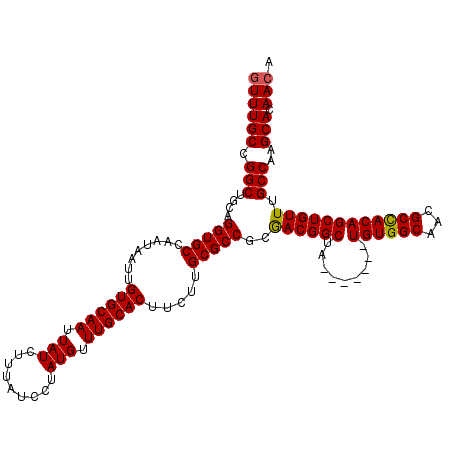
| Location | 7,543,516 – 7,543,627 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.55 |
| Mean single sequence MFE | -34.61 |
| Consensus MFE | -31.21 |
| Energy contribution | -31.25 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7543516 111 - 20766785 UGUUGUGCUUGGCAAACAGCUGUAGCGUUGCCACA-------UAGCCGUCGCGGCGCAAGAAGUGCAAACAUAGGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGGAGCCGGCAAAC .....((((.(((...(((.(((.(((((((.((.-------.....)).))))))).....((((((.................))))))........))).)))..)))))))... ( -33.43) >DroSec_CAF1 1762 111 - 1 UGUUGUGCUUGGCAAACAGCUGUGGCGUUGCCACA-------UAGCCGUCUCGGCGCAAGAAGUGCAAACAUAGGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGCAGCCGGCAAAC ((((((((...)))....((((..(.((.((((..-------..((((...)))).......((((((.................))))))......)))))))..)))))))))... ( -31.93) >DroSim_CAF1 1770 111 - 1 UGUUGUGCUUGGCAAACAGCUGUGGCGUUGCCACA-------UAGCCGUCGCGGCGCAAGAAGUGCAAACAUAGGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGCAGCCGGCAAAC .....((((.(((.....(((((((((..((....-------..)))))))))))(((....((((..............(((((((....))))))).)))).))).)))))))... ( -33.96) >DroEre_CAF1 4656 118 - 1 UGUUGUGCUUGGCAAACAGCUGUGGCGUUGCCACACUGCCAAUUGCCGUCGCGGCGCAAGAAGUGCAAACAUAAGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGCAGCCGGCAAAA .((((((..((((((.(((.((((((...)))))))))....)))))).))))))((.....((((((.................))))))......((((.......)))))).... ( -37.53) >DroYak_CAF1 1767 111 - 1 UGUUGUGCUUGGCAAACAGCUGUGGCGUUGCCACA-------UUGCCGUUGCGGCGCAAGAAGUGCAAGCAUAGGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGCAGCCGGCAAAC .(((((.........)))))((((((...))))))-------((((((((((.((((.....))))......(((.....(((((((....)))))))....))))))).)))))).. ( -36.20) >consensus UGUUGUGCUUGGCAAACAGCUGUGGCGUUGCCACA_______UAGCCGUCGCGGCGCAAGAAGUGCAAACAUAGGAUAAAGAUAAUUGCACAAUUAUUGGCACCUGCAGCCGGCAAAC ((((((((...)))....(((((((.((.((((...........((((...)))).......((((((.................))))))......))))))))))))))))))... (-31.21 = -31.25 + 0.04)



| Location | 7,543,554 – 7,543,664 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.06 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.34 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7543554 110 + 20766785 UUUAUCCUAUGUUUGCACUUCUUGCGCCGCGACGGCUA-------UGUGGCAACGCUACAGCUGUUUGCCAAGCACAACAAACAAAAUUAGAGGUGAGCGCGUGAUUAGCACUAACU ..........((((((...((.(((((((((((((((.-------.(((....)))...)))))))(((...)))..................))).))))).))...)))..))). ( -29.70) >DroSec_CAF1 1800 110 + 1 UUUAUCCUAUGUUUGCACUUCUUGCGCCGAGACGGCUA-------UGUGGCAACGCCACAGCUGUUUGCCAAGCACAACAAACAAAAUUAGAGGUGGGCGCGUGAUUAGCACUAACU ..........((((((...((.((((((.((((((((.-------.(((((...)))))))))))))......(((..(...........)..))))))))).))...)))..))). ( -32.70) >DroSim_CAF1 1808 110 + 1 UUUAUCCUAUGUUUGCACUUCUUGCGCCGCGACGGCUA-------UGUGGCAACGCCACAGCUGUUUGCCAAGCACAACAAACAAAAUUAGAGGUGGGCGCGUGAUUAGCACUAACU ..........((((((...((.(((((((((((((((.-------.(((((...)))))))))).))))....(((..(...........)..))))))))).))...)))..))). ( -33.60) >DroEre_CAF1 4694 116 + 1 UUUAUCUUAUGUUUGCACUUCUUGCGCCGCGACGGCAAUUGGCAGUGUGGCAACGCCACAGCUGUUUGCCAAGCACAACAAACAAAUGUGGAAGUGGGCGCGUGAUUAGGUCUAAC- ......((((((.(.((((((((((((((...))))..(((((((((((((...)))))).....))))))))))..(((......)))))))))).).))))))...........- ( -37.60) >DroYak_CAF1 1805 109 + 1 UUUAUCCUAUGCUUGCACUUCUUGCGCCGCAACGGCAA-------UGUGGCAACGCCACAGCUGUUUGCCAAGCACAACAAACAAAUGUGGAGGUGGGGUCGUGAUUAGGACUACU- ....(((((......(((.(((..(((((((.......-------))))))..(.(((((..((((((..........))))))..))))).))..)))..)))..))))).....- ( -34.70) >consensus UUUAUCCUAUGUUUGCACUUCUUGCGCCGCGACGGCUA_______UGUGGCAACGCCACAGCUGUUUGCCAAGCACAACAAACAAAAUUAGAGGUGGGCGCGUGAUUAGCACUAACU .......(((((.(.(((((((.((((((...)))).........((((((...))))))))((((((..........)))))).....))))))).).)))))............. (-29.70 = -29.34 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:32 2006