
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,530,192 – 7,530,294 |
| Length | 102 |
| Max. P | 0.797607 |

| Location | 7,530,192 – 7,530,294 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -12.36 |
| Energy contribution | -13.58 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7530192 102 + 20766785 GCAUUAACCAUAAUUUUGUUUAAUUAAAGUGGUUUAAACAAAUUAAAUGCGCUUAAGCUGCUUAAAUUACAGUGUUUAACGCAAAUGUAAUUUUGUAAAGCG--------- (((((.....((((((.((((((..........)))))))))))))))))((((....(((..(((((((((((.....)))...)))))))).))))))).--------- ( -19.60) >DroVir_CAF1 19050 102 + 1 GCAUUUACCAUAAUUGCGUUUAAUUACAGUGGUUUAAGCAAAUUAAAUGCGCUUAAGCGGCUUAAAUUACAGUGUUUAAGCUAAAUGCAAUUUUGUUAAGCG--------- ((..(((.((.((((((((((..........((((((((...........))))))))(((((((((......))))))))))))))))))).)).))))).--------- ( -29.50) >DroGri_CAF1 15487 102 + 1 GCAUUUACCAUAAUUGCGUUUAAUUAAAGUGUUUUAGGCAAAUUAAAUGCGCUUAAGCGGCUUAAAUUACAGUGUUUAAGCUAAAUGCAAUUUUGUUAAGAG--------- ...((((.((.((((((((((.....(((((((((((.....))))).))))))....(((((((((......))))))))))))))))))).)).))))..--------- ( -26.30) >DroMoj_CAF1 20214 102 + 1 GCAUUUAGCAUAAUUGCGUUUAAUUAAAGUAGUUUAAGCAAAUUAAAUGCGCUUAAGCGGCUCAAAUUACAGUGUUUAAGCUAAAUGCAAUUUUGUUAAGCG--------- ((..((((((.((((((((((..........((((((((...........))))))))((((.((((......)))).)))))))))))))).)))))))).--------- ( -30.00) >DroAna_CAF1 29495 96 + 1 ------ACUAUAAUUUCGCUUAAUUAAAGUGGUUUAAACAAAUUAAAUACGCUUAAGCUGCUUAAAUUACAGUAUUUAACGGAAAUGUAAUUUUGUUAAGCA--------- ------...........(((((((..(((..((((((.(...........).))))))..)))((((((((..............)))))))).))))))).--------- ( -18.54) >DroPer_CAF1 12512 104 + 1 GCACUAA-UAUAAUUGCAUUUAAUU-----GGUUU-AACAAAUUAAAUGCGCUUAAGCUGCACAAAUUACGGUGUUUAGGUUAAAUGUAAUUAUGUUGAGCAAAAAAAAGA ((..(((-(((((((((((((((((-----.....-((((...(((.(((((....)).)))....)))...))))..)))))))))))))))))))).)).......... ( -25.00) >consensus GCAUUUACCAUAAUUGCGUUUAAUUAAAGUGGUUUAAACAAAUUAAAUGCGCUUAAGCGGCUUAAAUUACAGUGUUUAAGCUAAAUGCAAUUUUGUUAAGCG_________ ........((.(((((((((((.........((((((.....))))))..((((((((.(((........)))))))))))))))))))))).))................ (-12.36 = -13.58 + 1.22)

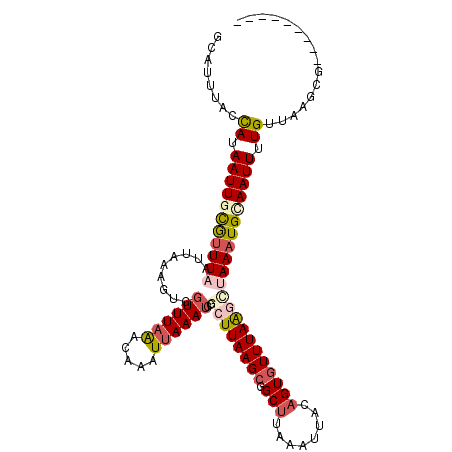

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:24 2006