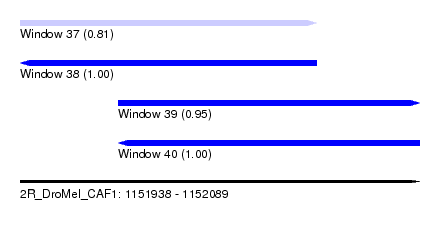
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,151,938 – 1,152,089 |
| Length | 151 |
| Max. P | 0.999971 |

| Location | 1,151,938 – 1,152,050 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.85 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -11.73 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1151938 112 + 20766785 UACAUUAGUCCACCGCAACUACAAGUACACCGCAUAC----ACUCACAGUAUUUCGUAGACUCGCGUCACAAAGCAUCAA----AAAAUGAUGCGGUGUUUGUUUACUCGUUUCCUUUGC ..............((((..((.((((....(((.((----(((...........((.(((....)))))...((((((.----....))))))))))).))).)))).)).....)))) ( -21.90) >DroSec_CAF1 2345 94 + 1 -----AAGUACACCGC--------AUAC-------------ACUCACAGUAUUUUGUAGACUUGCGUCACAAAGCACCGCAUCAUUUUUGAUGCGGUGUUUAUUCGCUCUUUUCAUUUGU -----((((..((...--------((((-------------.......))))...))..))))(((.....(((((((((((((....)))))))))))))...)))............. ( -27.70) >DroSim_CAF1 2992 107 + 1 -----AAGUACACCGC--------GUACACCGCAUACACUCACUCACAGUAUUUUGUAGACUUGCGUCACAAAGCACCACAUCAAAAAUGAAUCGGUGUUUAUUCGCUCUUUUCAUUUGU -----.........((--------(.((((((................((.((((((.(((....))))))))).))....(((....)))..)))))).....)))............. ( -18.80) >consensus _____AAGUACACCGC________GUACACCGCAUAC____ACUCACAGUAUUUUGUAGACUUGCGUCACAAAGCACCACAUCAAAAAUGAUGCGGUGUUUAUUCGCUCUUUUCAUUUGU ..........((((((.........................((.....)).((((((.(((....)))))))))..................))))))...................... (-11.73 = -12.40 + 0.67)

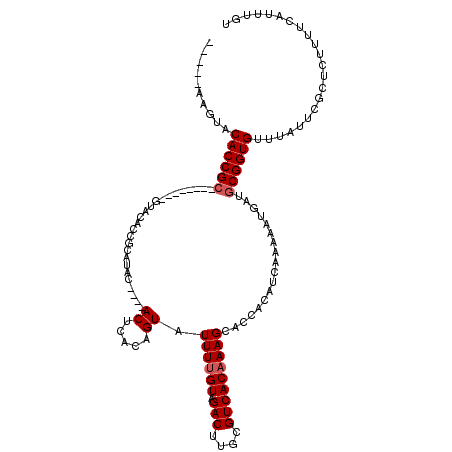
| Location | 1,151,938 – 1,152,050 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.85 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -18.72 |
| Energy contribution | -20.50 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.58 |
| SVM decision value | 5.05 |
| SVM RNA-class probability | 0.999971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1151938 112 - 20766785 GCAAAGGAAACGAGUAAACAAACACCGCAUCAUUUU----UUGAUGCUUUGUGACGCGAGUCUACGAAAUACUGUGAGU----GUAUGCGGUGUACUUGUAGUUGCGGUGGACUAAUGUA ((((.....((((((......((((((((((((((.----..(.((.((((((((....))).))))).)))...))))----).)))))))))))))))..)))).............. ( -34.90) >DroSec_CAF1 2345 94 - 1 ACAAAUGAAAAGAGCGAAUAAACACCGCAUCAAAAAUGAUGCGGUGCUUUGUGACGCAAGUCUACAAAAUACUGUGAGU-------------GUAU--------GCGGUGUACUU----- ......................((((((((((....)))))))))).((((((((....))).)))))(((((((....-------------....--------)))))))....----- ( -31.90) >DroSim_CAF1 2992 107 - 1 ACAAAUGAAAAGAGCGAAUAAACACCGAUUCAUUUUUGAUGUGGUGCUUUGUGACGCAAGUCUACAAAAUACUGUGAGUGAGUGUAUGCGGUGUAC--------GCGGUGUACUU----- .....................(((((.((((((((.....(..(((.((((((((....))).))))).)))..)))))))))....(((.....)--------)))))))....----- ( -30.60) >consensus ACAAAUGAAAAGAGCGAAUAAACACCGCAUCAUUUUUGAUGUGGUGCUUUGUGACGCAAGUCUACAAAAUACUGUGAGU____GUAUGCGGUGUAC________GCGGUGUACUU_____ ......................((((((...................((((((((....))).)))))(((((((............)))))))..........)))))).......... (-18.72 = -20.50 + 1.78)

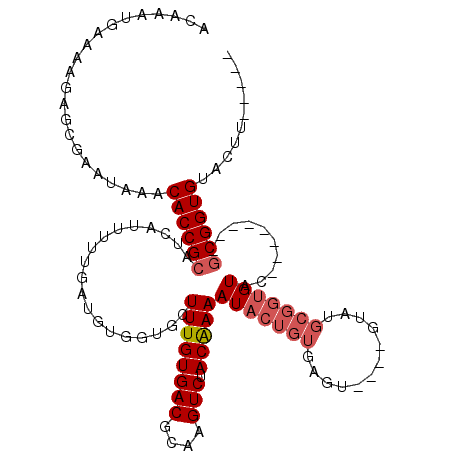
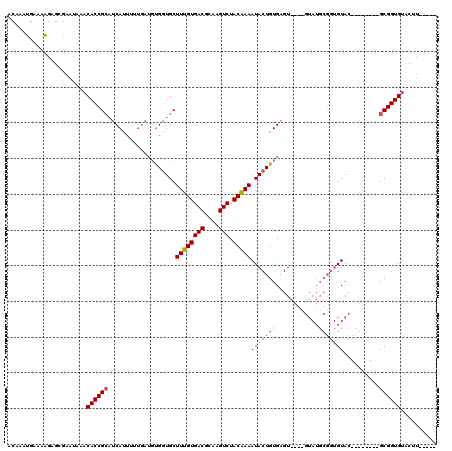
| Location | 1,151,975 – 1,152,089 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -18.26 |
| Energy contribution | -19.60 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1151975 114 + 20766785 -ACUCACAGUAUUUCGUAGACUCGCGUCACAAAGCAUCAA----AAAAUGAUGCGGUGUUUGUUUACUCGUUUCCUUUGCGCACUCCUCCUAUGCGGUGCCAAAUAUGUGCAGGUACUU -.((((((.......((((((......(((...((((((.----....)))))).)))...))))))........((((.(((((.(......).)))))))))..)))).))...... ( -30.00) >DroSec_CAF1 2360 111 + 1 -ACUCACAGUAUUUUGUAGACUUGCGUCACAAAGCACCGCAUCAUUUUUGAUGCGGUGUUUAUUCGCUCUUUUCAUUUGUGCACUCCUUCUAUGCGGUGCGAUAUAUGAGCA------- -.............(((.(((....))))))(((((((((((((....)))))))))))))....((((..........((((((.(......).))))))......)))).------- ( -37.29) >DroSim_CAF1 3019 112 + 1 CACUCACAGUAUUUUGUAGACUUGCGUCACAAAGCACCACAUCAAAAAUGAAUCGGUGUUUAUUCGCUCUUUUCAUUUGUGCACUUCCUCUAUGCGGUGCUAUAUAUGAGCA------- ..((((.......((((.(((....)))))))((((((.......(((((((..((((......))))...)))))))..(((.........))))))))).....))))..------- ( -27.40) >consensus _ACUCACAGUAUUUUGUAGACUUGCGUCACAAAGCACCACAUCAAAAAUGAUGCGGUGUUUAUUCGCUCUUUUCAUUUGUGCACUCCUUCUAUGCGGUGCCAUAUAUGAGCA_______ ..((((......(((((.(((....))))))))(((((.......((((((...((((......))))....))))))..(((.........))))))))......))))......... (-18.26 = -19.60 + 1.34)

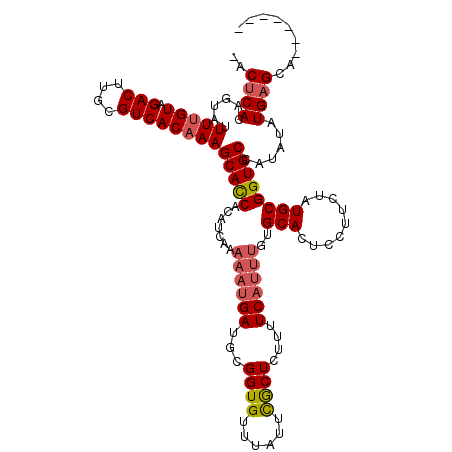
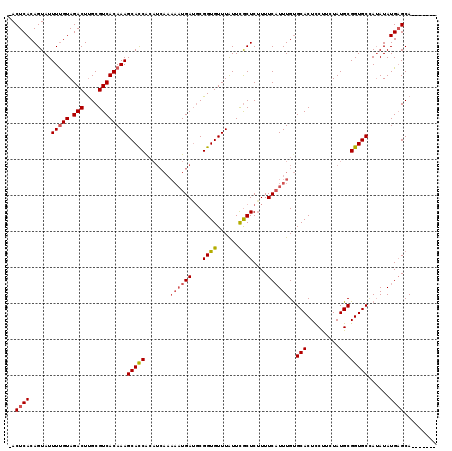
| Location | 1,151,975 – 1,152,089 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -24.71 |
| Energy contribution | -25.60 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.70 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1151975 114 - 20766785 AAGUACCUGCACAUAUUUGGCACCGCAUAGGAGGAGUGCGCAAAGGAAACGAGUAAACAAACACCGCAUCAUUUU----UUGAUGCUUUGUGACGCGAGUCUACGAAAUACUGUGAGU- ......((.(((...((((((((..(......)..)))).))))(....).((((..........((((((....----.))))))((((((((....))).))))).))))))))).- ( -29.70) >DroSec_CAF1 2360 111 - 1 -------UGCUCAUAUAUCGCACCGCAUAGAAGGAGUGCACAAAUGAAAAGAGCGAAUAAACACCGCAUCAAAAAUGAUGCGGUGCUUUGUGACGCAAGUCUACAAAAUACUGUGAGU- -------.(((((((..((((...((((.......)))).....(....)..)))).....((((((((((....)))))))))).((((((((....))).)))))....)))))))- ( -39.60) >DroSim_CAF1 3019 112 - 1 -------UGCUCAUAUAUAGCACCGCAUAGAGGAAGUGCACAAAUGAAAAGAGCGAAUAAACACCGAUUCAUUUUUGAUGUGGUGCUUUGUGACGCAAGUCUACAAAAUACUGUGAGUG -------.((((((((((.(((((((((...((((((((.............))((((........)))))))))).)))))))))((((((((....))).)))))))).))))))). ( -35.92) >consensus _______UGCUCAUAUAUAGCACCGCAUAGAAGGAGUGCACAAAUGAAAAGAGCGAAUAAACACCGCAUCAUUUUUGAUGUGGUGCUUUGUGACGCAAGUCUACAAAAUACUGUGAGU_ ........(((((((....(((((((((.......))))...........(((((.........))).))...........)))))((((((((....))).)))))....))))))). (-24.71 = -25.60 + 0.89)

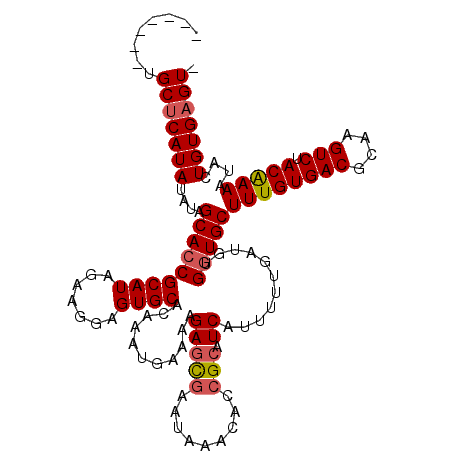
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:41 2006