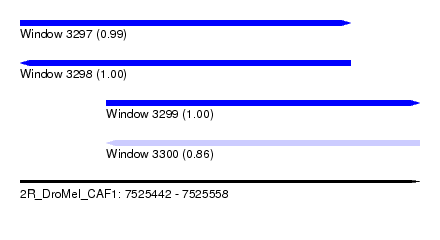
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,525,442 – 7,525,558 |
| Length | 116 |
| Max. P | 1.000000 |

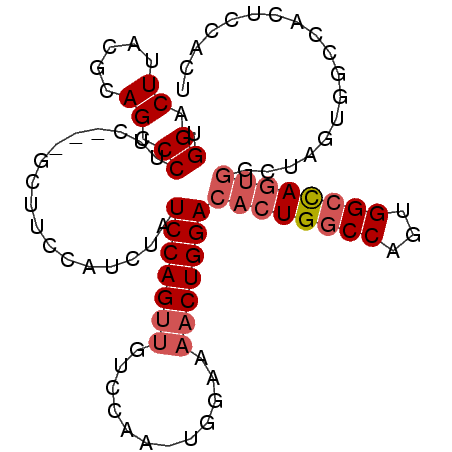
| Location | 7,525,442 – 7,525,538 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 67.36 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -12.69 |
| Energy contribution | -14.47 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
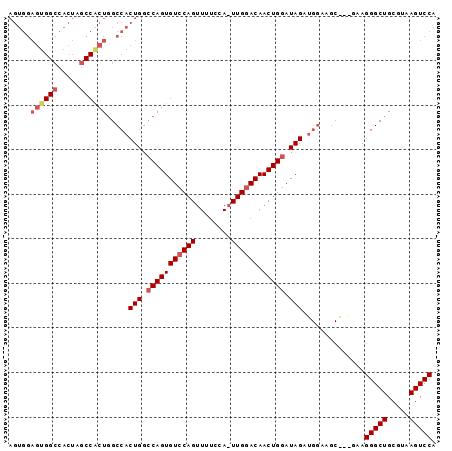
>2R_DroMel_CAF1 7525442 96 + 20766785 UGGACUUAUGCAGCCCUUCUGCGCUUCCAUCUAUCCAGUUGUCCAACUGGAAAACUGGACACUGGCCAGUGGCUAGCGGCUAGUGGCCACUCCACU ((((....(((((.....)))))..))))....(((((((.(((....))).)))))))...((((((.((((.....)))).))))))....... ( -38.30) >DroSec_CAF1 7311 84 + 1 UGGACUUACGCAGCCCUU------------CUAUCCAGUUGUGCAAGUGGAAAACUGGACACUGUCCAGUGGCCAGUGGCUAGUGGCCACUCCACU (((((((.((((((....------------.......)))))).))))((...(((((((...)))))))..))((((((.....))))))))).. ( -31.80) >DroYak_CAF1 7437 74 + 1 UGGACUUACGCAGCCCUUCCCUGCUUCCAUCUAUCCAGUUGUCCAA--------CUGGACACUGGCCAGUGGCCACU----CCACU---------- ((((.....((((.......))))..(((.((..(((((.((((..--------..)))))))))..)))))....)----)))..---------- ( -23.80) >consensus UGGACUUACGCAGCCCUUC___GCUUCCAUCUAUCCAGUUGUCCAA_UGGAAAACUGGACACUGGCCAGUGGCCAGUGGCUAGUGGCCACUCCACU .((.((.....)).)).................(((((((............)))))))((((((((...)))))))).................. (-12.69 = -14.47 + 1.78)

| Location | 7,525,442 – 7,525,538 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 67.36 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -22.21 |
| Energy contribution | -24.27 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.64 |
| SVM decision value | 7.23 |
| SVM RNA-class probability | 1.000000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
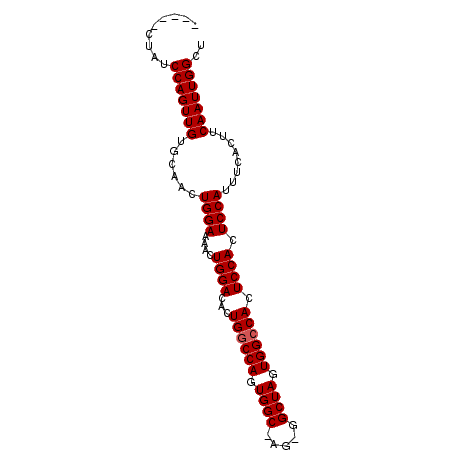
>2R_DroMel_CAF1 7525442 96 - 20766785 AGUGGAGUGGCCACUAGCCGCUAGCCACUGGCCAGUGUCCAGUUUUCCAGUUGGACAACUGGAUAGAUGGAAGCGCAGAAGGGCUGCAUAAGUCCA (((((((((((.....))))))..)))))..(((.(((((((((.(((....))).)))))))))..)))..........(((((.....))))). ( -41.80) >DroSec_CAF1 7311 84 - 1 AGUGGAGUGGCCACUAGCCACUGGCCACUGGACAGUGUCCAGUUUUCCACUUGCACAACUGGAUAG------------AAGGGCUGCGUAAGUCCA .((..((((((((........))))))))..))..(((((((((...(....)...))))))))).------------..(((((.....))))). ( -32.20) >DroYak_CAF1 7437 74 - 1 ----------AGUGG----AGUGGCCACUGGCCAGUGUCCAG--------UUGGACAACUGGAUAGAUGGAAGCAGGGAAGGGCUGCGUAAGUCCA ----------.....----.((..((((((.(((((((((..--------..)))).))))).))).)))..))......(((((.....))))). ( -29.70) >consensus AGUGGAGUGGCCACUAGCCACUGGCCACUGGCCAGUGUCCAGUUUUCCA_UUGGACAACUGGAUAGAUGGAAGC___GAAGGGCUGCGUAAGUCCA .....((((((.....)))))).....(((.(((((((((((........)))))).))))).)))..............(((((.....))))). (-22.21 = -24.27 + 2.06)

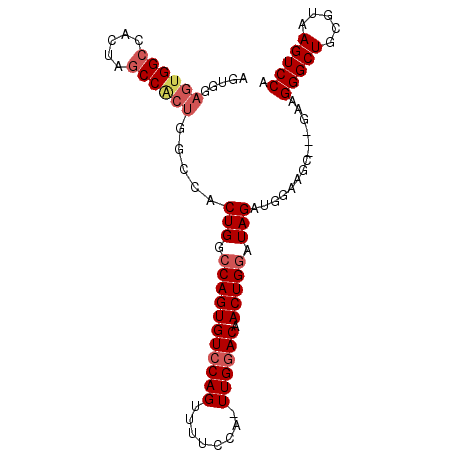
| Location | 7,525,467 – 7,525,558 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 82.84 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -25.14 |
| Energy contribution | -25.81 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
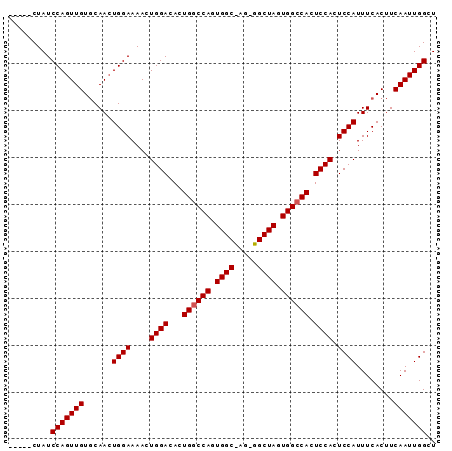
>2R_DroMel_CAF1 7525467 91 + 20766785 UCCAUCUAUCCAGUUGUCCAACUGGAAAACUGGACACUGGCCAGUGGCUAGCGGCUAGUGGCCACUCCACUCCAUUUCACUUCAAUUGGCU .(((....(((((((.(((....))).)))))))...((((((.((((.....)))).))))))......................))).. ( -32.50) >DroSec_CAF1 7329 86 + 1 -----CUAUCCAGUUGUGCAAGUGGAAAACUGGACACUGUCCAGUGGCCAGUGGCUAGUGGCCACUCCACUCCAUUUCACUUCAAUUGGCU -----....(((((((((.(((((((..(((((((...)))))))((..((((((.....))))))))..))))))))))...)))))).. ( -34.60) >DroSim_CAF1 7290 72 + 1 -----CUAUCCAGUUGUGCAACUGGAAAACUGGACACUGGCCA--------------GUGGCCACUCCACUCCAUUUCACUUCAAUUGGCU -----....(((((((((.((.((((....((((...(((((.--------------..))))).)))).)))).)))))...)))))).. ( -20.40) >consensus _____CUAUCCAGUUGUGCAACUGGAAAACUGGACACUGGCCAGUGGC_AG_GGCUAGUGGCCACUCCACUCCAUUUCACUUCAAUUGGCU .........(((((((......((((....((((...((((((.((((.....)))).)))))).)))).))))........))))))).. (-25.14 = -25.81 + 0.67)

| Location | 7,525,467 – 7,525,558 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 82.84 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -19.50 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
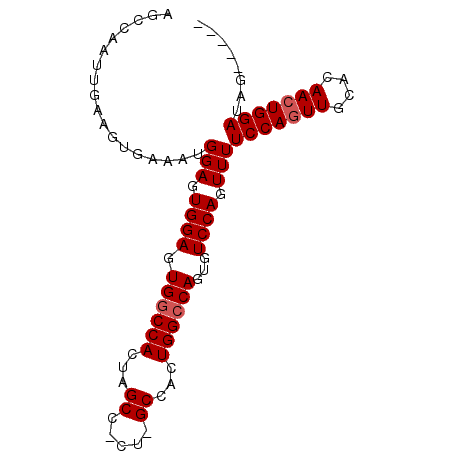
>2R_DroMel_CAF1 7525467 91 - 20766785 AGCCAAUUGAAGUGAAAUGGAGUGGAGUGGCCACUAGCCGCUAGCCACUGGCCAGUGUCCAGUUUUCCAGUUGGACAACUGGAUAGAUGGA ..(((.((......)).)))(((((((((((.....))))))..)))))..(((.(((((((((.(((....))).)))))))))..))). ( -36.30) >DroSec_CAF1 7329 86 - 1 AGCCAAUUGAAGUGAAAUGGAGUGGAGUGGCCACUAGCCACUGGCCACUGGACAGUGUCCAGUUUUCCACUUGCACAACUGGAUAG----- ..(((...(..(((.....((((((((.(((((........)))))(((((((...))))))).)))))))).)))..))))....----- ( -29.80) >DroSim_CAF1 7290 72 - 1 AGCCAAUUGAAGUGAAAUGGAGUGGAGUGGCCAC--------------UGGCCAGUGUCCAGUUUUCCAGUUGCACAACUGGAUAG----- ..................(((.((((.(((((..--------------.)))))...)))).)))(((((((....)))))))...----- ( -21.00) >consensus AGCCAAUUGAAGUGAAAUGGAGUGGAGUGGCCACUAGCC_CU_GCCACUGGCCAGUGUCCAGUUUUCCAGUUGCACAACUGGAUAG_____ ..................(((.((((.((((((...((.....))...))))))...)))).)))(((((((....)))))))........ (-19.50 = -20.33 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:22 2006