| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,522,885 – 7,522,979 |
| Length | 94 |
| Max. P | 0.955738 |

| Location | 7,522,885 – 7,522,979 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 95.02 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -19.21 |
| Energy contribution | -19.21 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7522885 94 + 20766785 UAUGGCGCAAAAGAAAAGCGCAUAAAA-AAUGGGG----GGGAAAAAAACUGCAGUCCAAAAGAAGAAUGCGCUGCUCUCUAAACUCUCUGCCGUGGCA ....((((.........))))......-.((((.(----((((........(((((.((.........)).))))).........))))).)))).... ( -23.13) >DroSec_CAF1 4774 95 + 1 UAUGGCGCAAAAGAAAAGCGCAUAAAAAAAUGGGG----GGAAAAAAAACUGCAGUCCAAAAGAAGAAUGCGCUGCUCUCUAAACUCUCUGCCGUGGCA ....((((.........))))........((((((----(((.........(((((.((.........)).))))).........))))).)))).... ( -20.27) >DroSim_CAF1 4745 99 + 1 UAUGGCGCAAAAGAAAAGCGCAUAAAAAAAUGGGGGGGGGGGAAAAAAACUGCAGUCCAAAAGAAGAAUGCGCUGCUCUCUAAACUCUCUGCCGUGGCA ....((((.........))))........((((.((((((...........(((((.((.........)).)))))........)))))).)))).... ( -25.01) >DroEre_CAF1 4823 94 + 1 UAUGGCGCAAAAGAAAAGCGCAUAAAA-AAUGGGG----GGAAAAAAAACUGCAGUCCAAAAGAAGAAUGCGCUGCUCUCUAAACUCUCAGCCAUAGCA ((((((((.........))........-..(((((----(...........(((((.((.........)).)))))........))))))))))))... ( -21.31) >DroYak_CAF1 4823 92 + 1 UAUGGCGCAAAAGAAAAGCGCAUAAAA-AAUGGGG----G--AAAAAAACUGCAGUCCAAAAGAAGAAUGCGCUGCUCUCUAAACUCUCUGCCAUGGCA (((((((....(((..(((((((....-..((.((----.--.......)).)).((.....))...)))))))..)))..........)))))))... ( -19.54) >consensus UAUGGCGCAAAAGAAAAGCGCAUAAAA_AAUGGGG____GGAAAAAAAACUGCAGUCCAAAAGAAGAAUGCGCUGCUCUCUAAACUCUCUGCCGUGGCA (((((((....(((..(((((((.........(((....((........))....))).........)))))))..)))..........)))))))... (-19.21 = -19.21 + 0.00)


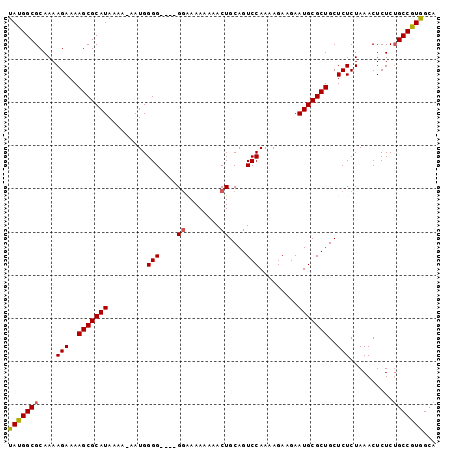
| Location | 7,522,885 – 7,522,979 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 95.02 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -20.24 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7522885 94 - 20766785 UGCCACGGCAGAGAGUUUAGAGAGCAGCGCAUUCUUCUUUUGGACUGCAGUUUUUUUCCC----CCCCAUU-UUUUAUGCGCUUUUCUUUUGCGCCAUA (((....)))....((..((((((.(((((((........(((.................----..)))..-....))))))).)))))).))...... ( -20.45) >DroSec_CAF1 4774 95 - 1 UGCCACGGCAGAGAGUUUAGAGAGCAGCGCAUUCUUCUUUUGGACUGCAGUUUUUUUUCC----CCCCAUUUUUUUAUGCGCUUUUCUUUUGCGCCAUA (((....)))....((..((((((.(((((((........(((.................----..))).......))))))).)))))).))...... ( -20.87) >DroSim_CAF1 4745 99 - 1 UGCCACGGCAGAGAGUUUAGAGAGCAGCGCAUUCUUCUUUUGGACUGCAGUUUUUUUCCCCCCCCCCCAUUUUUUUAUGCGCUUUUCUUUUGCGCCAUA (((....)))....((..((((((.(((((((........(((.......................))).......))))))).)))))).))...... ( -20.66) >DroEre_CAF1 4823 94 - 1 UGCUAUGGCUGAGAGUUUAGAGAGCAGCGCAUUCUUCUUUUGGACUGCAGUUUUUUUUCC----CCCCAUU-UUUUAUGCGCUUUUCUUUUGCGCCAUA ...((((((.....((..((((((.(((((((........(((.................----..)))..-....))))))).)))))).)))))))) ( -22.25) >DroYak_CAF1 4823 92 - 1 UGCCAUGGCAGAGAGUUUAGAGAGCAGCGCAUUCUUCUUUUGGACUGCAGUUUUUUU--C----CCCCAUU-UUUUAUGCGCUUUUCUUUUGCGCCAUA ....(((((.....((..((((((.(((((((........(((..............--.----..)))..-....))))))).)))))).))))))). ( -21.97) >consensus UGCCACGGCAGAGAGUUUAGAGAGCAGCGCAUUCUUCUUUUGGACUGCAGUUUUUUUCCC____CCCCAUU_UUUUAUGCGCUUUUCUUUUGCGCCAUA ......(((.....((..((((((.(((((((........(((.......................))).......))))))).)))))).)))))... (-20.24 = -20.24 + 0.00)

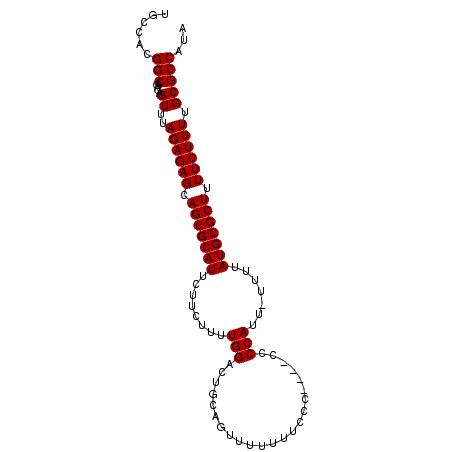
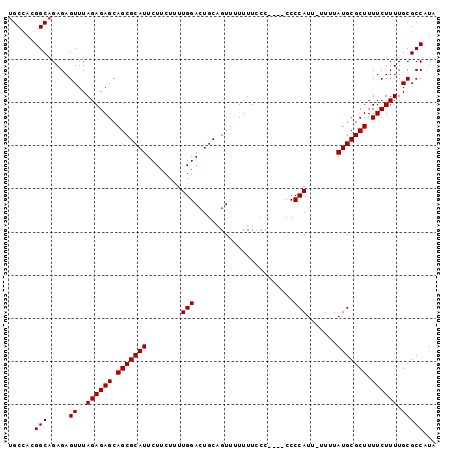
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:18 2006