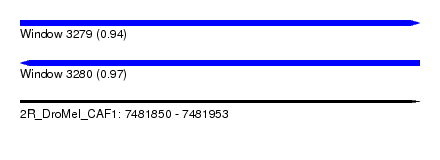
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,481,850 – 7,481,953 |
| Length | 103 |
| Max. P | 0.965195 |

| Location | 7,481,850 – 7,481,953 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.47 |
| Mean single sequence MFE | -45.65 |
| Consensus MFE | -34.53 |
| Energy contribution | -34.65 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7481850 103 + 20766785 ------------GGCGCAACAGGAGUGGCUGGGGGAAAGGCCAGAGGAGGCGGUGGGCCAACGAUGGCCCCACCACUGGCCACCACUCCAGCCGCAAACAAUGCAACUGCGGUUA ------------..((((........((((((((....((((((.(....)(((((((((....))).)))))).))))))....))))))))(((.....)))...)))).... ( -50.50) >DroSec_CAF1 98208 103 + 1 ------------AGCGCAACAGGAGUGGCUGGGGGAAAGGCCCGAGGAGGCGGUGUGGCAACGAUGGCCCCACCACUGGCCACCAGUCCAGCCGCAAACAAUGCAACUGCGGUUA ------------..((((........(((((((((.....)))..((.(((((((.(((.......))).))))....))).))...))))))(((.....)))...)))).... ( -41.30) >DroSim_CAF1 90554 103 + 1 ------------AGCGCAACAGGAGUGGCUGGGGGAAAGGCCCGAGGAGGCGGUGGGCCAACGAUGGCCCCACCACUGGCCACCAGUCUAGCCGCAAACAAUGCAACUGCGGUUA ------------..((((...((...((((((.((..(((((......)))(((((((((....))).)))))).))..)).))))))...))(((.....)))...)))).... ( -46.30) >DroYak_CAF1 96568 115 + 1 GGUUCGGGACCAGGAGCAACAGGAGUUGCUGGGGGAAAGAUCAGAGGAGGCAGUGGGCCAACAAUGUCCCCAGCACUGGCCGCCAGCCCAGCGGCAAACAAUGCAACUGCGGCCA (((...((.((((.((((((....)))))).(((((.....((..(..(((.....)))..)..)))))))....))))))))).((((((..(((.....)))..))).))).. ( -44.50) >consensus ____________AGCGCAACAGGAGUGGCUGGGGGAAAGGCCAGAGGAGGCGGUGGGCCAACGAUGGCCCCACCACUGGCCACCAGUCCAGCCGCAAACAAUGCAACUGCGGUUA ...............((..(((....(((((((((...((((((.((.(((.((.(.....).)).)))))....)))))).))..)))))))(((.....)))..)))..)).. (-34.53 = -34.65 + 0.12)

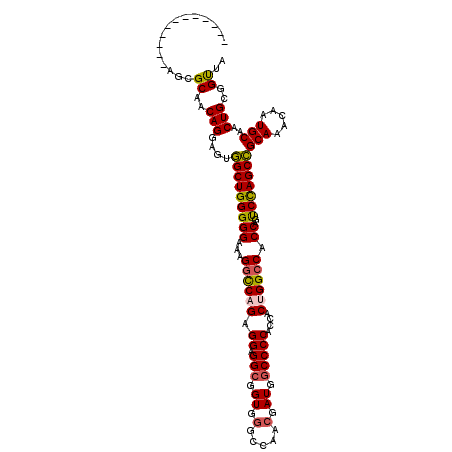
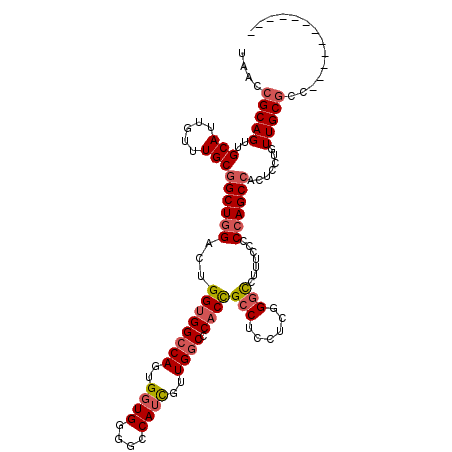
| Location | 7,481,850 – 7,481,953 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.47 |
| Mean single sequence MFE | -46.55 |
| Consensus MFE | -34.76 |
| Energy contribution | -35.95 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7481850 103 - 20766785 UAACCGCAGUUGCAUUGUUUGCGGCUGGAGUGGUGGCCAGUGGUGGGGCCAUCGUUGGCCCACCGCCUCCUCUGGCCUUUCCCCCAGCCACUCCUGUUGCGCC------------ ....((((((((((.....)))))))((((((((((((((.((((((((((....)))))..)))))....)))))).........))))))))....)))..------------ ( -53.00) >DroSec_CAF1 98208 103 - 1 UAACCGCAGUUGCAUUGUUUGCGGCUGGACUGGUGGCCAGUGGUGGGGCCAUCGUUGCCACACCGCCUCCUCGGGCCUUUCCCCCAGCCACUCCUGUUGCGCU------------ ....(((((..(((.....)))((((((...((.((((.((((((.(((.......))).)))))).......))))...)).)))))).......)))))..------------ ( -41.60) >DroSim_CAF1 90554 103 - 1 UAACCGCAGUUGCAUUGUUUGCGGCUAGACUGGUGGCCAGUGGUGGGGCCAUCGUUGGCCCACCGCCUCCUCGGGCCUUUCCCCCAGCCACUCCUGUUGCGCU------------ ....((((((((((.....))))))).(((.((((((..((((((((.(((....)))))))))))......(((.......))).))))))...))))))..------------ ( -45.40) >DroYak_CAF1 96568 115 - 1 UGGCCGCAGUUGCAUUGUUUGCCGCUGGGCUGGCGGCCAGUGCUGGGGACAUUGUUGGCCCACUGCCUCCUCUGAUCUUUCCCCCAGCAACUCCUGUUGCUCCUGGUCCCGAACC .((((.((((.(((.....))).))))))))((.((((((....(((((((..(..(((.....)))..)..)).....))))).((((((....)))))).))))))))..... ( -46.20) >consensus UAACCGCAGUUGCAUUGUUUGCGGCUGGACUGGUGGCCAGUGGUGGGGCCAUCGUUGGCCCACCGCCUCCUCGGGCCUUUCCCCCAGCCACUCCUGUUGCGCC____________ ....(((((..(((.....)))((((((...((((((((..((((....))))..)))).))))(((......))).......)))))).......))))).............. (-34.76 = -35.95 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:03 2006