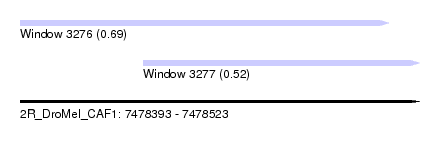
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,478,393 – 7,478,523 |
| Length | 130 |
| Max. P | 0.687553 |

| Location | 7,478,393 – 7,478,513 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -29.68 |
| Energy contribution | -28.02 |
| Covariance contribution | -1.66 |
| Combinations/Pair | 1.42 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
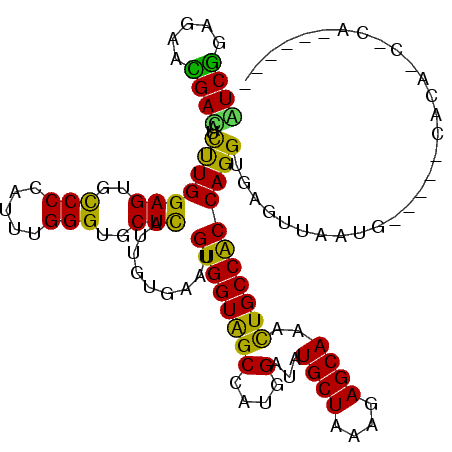
>2R_DroMel_CAF1 7478393 120 + 20766785 GUCGGAGAACGACAUCUUGGAGUGCCCCAUCUGGGUGCUCAUCGUUAAUGUGGUGGCCAUGGAUAUGCUGAAGAGCAAACUGCCACCAGGUGGGUUCAUCAAAGCCAUGUCCCAUGCAGA ((((.....))))......(((..(((.....)))..)))........(((((((((...((...((((....))))..))))))))..(((((..(((.......))).)))))))).. ( -40.40) >DroPse_CAF1 119495 109 + 1 AUCGGAGAGCGAUGUCUUGGAGUGCCCCAUUUGGGUGCUCAUUGUGAAUGUGGUAGCCAUGGAUAUGCUCAAGAGCAAGCUGCCACCAGGUGAGUUAAUG-----CACAGCACA------ .((.((((......)))).))(((((((....))).(((((((......((((((((........((((....)))).))))))))..))))))).....-----....)))).------ ( -34.90) >DroGri_CAF1 96531 105 + 1 AUCGGAGAAUGACAUUUUGGAGUGCCCCAUUUGGGUGCUCAUUGUGAAUGUGGUAGCCAUGGAUAUGCUAAAGAGCAAACUGCCGCCAGGUGAGUU-AUG-----UGCA---------AG ...((...((.((((((..(((..(((.....)))..))).....)))))).))..)).((.(((((((....))).(((((((....))).))))-)))-----).))---------.. ( -31.90) >DroWil_CAF1 125933 114 + 1 GUCAGAGAAUGACAUUUUGGAGUGCCCCAUUUGGGUGCUUAUUGUUAAUGUGGUAGCCAUGGAUAUGCUAAAGAGCAAACUGCCACCAGGUGAGACAAUUGGUUGUUUGUCGUU------ ((((.....))))..(((((((..(((.....)))..))).........((((((((....)...((((....))))..)))))))))))...(((((........)))))...------ ( -32.20) >DroAna_CAF1 90861 104 + 1 GUCGGAGAACGAUGUCCUGGAGUGUCCCAUUUGGGUGCUCAUUGUCAAUGUGGUGGCCAUGGAUAUGCUAAAGAGCAAACUGCCACCAGGUAGGCUCUACACA-C--------------- ((((.....))))......(((((.(((....))))))))...(((.((.(((((((...((...((((....))))..))))))))).)).)))........-.--------------- ( -33.40) >DroPer_CAF1 89380 109 + 1 AUCGGAGAGCGAUGUCUUGGAGUGCCCCAUUUGGGUGCUCAUUGUGAAUGUGGUAGCCAUGGAUAUGCUCAAAAGCAAGUUGCCACCAGGUGAGUUAAUG-----CACAGCACA------ ......((((.((((((..(((..(((.....)))..))).........((((...))))))))))))))....((((.(..((....))..).)...))-----)........------ ( -32.40) >consensus AUCGGAGAACGACAUCUUGGAGUGCCCCAUUUGGGUGCUCAUUGUGAAUGUGGUAGCCAUGGAUAUGCUAAAGAGCAAACUGCCACCAGGUGAGUUAAUG_____CACA_C_CA______ ((((.....))))..(((((((..(((.....)))..))).........((((((((....)...((((....))))..))))))))))).............................. (-29.68 = -28.02 + -1.66)

| Location | 7,478,433 – 7,478,523 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.11 |
| Mean single sequence MFE | -22.37 |
| Consensus MFE | -15.78 |
| Energy contribution | -15.70 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7478433 90 + 20766785 AUCGUUAAUGUGGUGGCCAUGGAUAUGCUGAAGAGCAAACUGCCACCAGGUGGGUUCAUCAAAGCCAUGUCCCAUGCAGAUUUG----------------AGUCU---U .(((....(((((((((...((...((((....))))..))))))))..(((((..(((.......))).))))))))....))----------------)....---. ( -23.90) >DroPse_CAF1 119535 70 + 1 AUUGUGAAUGUGGUAGCCAUGGAUAUGCUCAAGAGCAAGCUGCCACCAGGUGAGUUAAUG-----CACAGCACA----------------------------------C ..((((...((((((((........((((....)))).))))))))...(((........-----)))..))))----------------------------------. ( -19.50) >DroSec_CAF1 94845 75 + 1 AUCGUCAAUGUGGUGGCCAUGGAUAUGCUGAAGAGCAAGCUGCCACCAGGUGGGUUCAUCGAAGCCAUAUCCCA----------------------------------U .......(((((((..(.((((((.((((....))))..(..((....))..))))))).)..)))))))....----------------------------------. ( -22.30) >DroSim_CAF1 87181 75 + 1 AUCGUCAAUGUGGUGGCCAUGGAUAUGCUGAAGAGCAAGCUGCCACCAGGUGGGUUCAUCGAAGCCAUAUCCCA----------------------------------C .......(((((((..(.((((((.((((....))))..(..((....))..))))))).)..)))))))....----------------------------------. ( -22.30) >DroYak_CAF1 93241 108 + 1 AUCGUCAACGUGGUGGCCAUGGAUAUGCUGAAGAGCAAGCUGCCGCCAGGUGGGUUCAUCGAAGCCAUAUCACGUGCAGAUUUGGCUGCUAGUCUUUUUUUGUUUUUU- ..........(((..((((......((((....))))..((((((....((((.(((...))).))))....)).))))...))))..))).................- ( -30.10) >DroPer_CAF1 89420 70 + 1 AUUGUGAAUGUGGUAGCCAUGGAUAUGCUCAAAAGCAAGUUGCCACCAGGUGAGUUAAUG-----CACAGCACA----------------------------------C ..((((...((((((((........((((....)))).))))))))...(((........-----)))..))))----------------------------------. ( -16.10) >consensus AUCGUCAAUGUGGUGGCCAUGGAUAUGCUGAAGAGCAAGCUGCCACCAGGUGGGUUCAUCGAAGCCAUAUCACA__________________________________C .......((.(((((((...((...((((....))))..))))))))).)).......................................................... (-15.78 = -15.70 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:00 2006