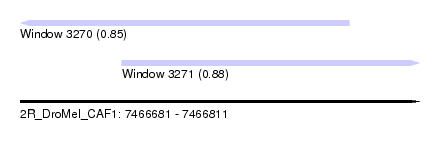
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,466,681 – 7,466,811 |
| Length | 130 |
| Max. P | 0.876800 |

| Location | 7,466,681 – 7,466,788 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.45 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -11.00 |
| Energy contribution | -11.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7466681 107 - 20766785 UUGGCCACACUCAGCUGA----CUUACUGCCUUGGCGGCCUGUAGGCAAAUUAAAAAUGCACAACGCAAUGCGAUGGCCUAACGGUAAUUAAUGCAAUUGGUCAAUACAAC------- (((((((......(((.(----(...((((....))))...)).)))..........((((((.(((...))).))(((....)))......))))..)))))))......------- ( -29.90) >DroVir_CAF1 89488 105 - 1 --GUUUUUGUUGCGGCA--------ACUUAAA--AAUGGCCAUAGGCAAAUUAAAAAUGCACAACGCAAUGCGAUGGCCUAACGGUAAUUAAUGCAAUUGGUCAAUACAACAA-CAAC --(((.((((((.(...--------.).....--..((((((...(((.............((.(((...))).))(((....)))......)))...))))))...))))))-.))) ( -26.50) >DroGri_CAF1 84198 105 - 1 --GUUGUUGUUGAGGCA--------ACUUCCA--AAUGGCGAUAGCAAAAUUAAAAAUGCACAACGCAAUGCGAUGGCCUAACGGUAAUUAAUGCAAUUGGUCAAUACAACAG-CACC --((((((((.(((...--------.)))(((--(...((....))...........((((((.(((...))).))(((....)))......)))).)))).....)))))))-)... ( -27.70) >DroMoj_CAF1 107155 99 - 1 --------GUUGCGGCA--------ACUGCCA--AAUGGCCAUAGGCAAAUUAAAAAUGCACAACGCAAUGCGAUGGCCUAACGGUAAUUAAUGCAAUUGGUCAAUACAACAA-CAGC --------((((.(((.--------...))).--..((((((...(((.............((.(((...))).))(((....)))......)))...))))))...))))..-.... ( -27.90) >DroAna_CAF1 80051 88 - 1 -----------------------UUACAGCCCGGUCUCCUCGUAGGCAAAUUAAAAAUGCACAACGCAAUGCGAUGGCCUAACGGUAAUUAAUGCAAUUGGUCAAUACAAC------- -----------------------.....(.(((((((......))))..........((((((.(((...))).))(((....)))......))))..))).)........------- ( -15.50) >DroPer_CAF1 78028 112 - 1 UUCGCC---UUUAGCUGUGCUUUUUACUGCCUUG---GCCCAUAGGCAAAUUAAAAAUGCACAACGCAAUGCGAUGGCCUAACGGUAAUUAAUGCAAUUGGUCAAUACAAAAAACACC .((((.---....(.(((((((((((.(((((..---......)))))...)))))).))))).).....))))(((((.....(((.....)))....))))).............. ( -28.10) >consensus __G_____GUUGAGGCA________ACUGCCA__AAUGCCCAUAGGCAAAUUAAAAAUGCACAACGCAAUGCGAUGGCCUAACGGUAAUUAAUGCAAUUGGUCAAUACAACAA_CA_C .........................................................((((((.(((...))).))(((....)))......))))...................... (-11.00 = -11.00 + 0.00)


| Location | 7,466,714 – 7,466,811 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.92 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
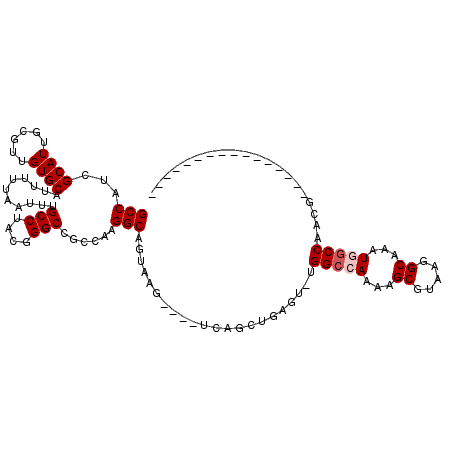
>2R_DroMel_CAF1 7466714 97 + 20766785 GCCAUCGCAUUGCGUUGUGCAUUUUUAAUUUGCCUACAGGCCGCCAAGGCAGUAAG----UCAGCUGAGUGUGGCCAAAAGCGUAAGGCAAAUGGCCAACG---------------- (((((.((.(((((((..(((.........))).....((((((.....((((...----...))))...))))))...))))))).))..))))).....---------------- ( -32.10) >DroPse_CAF1 108201 111 + 1 GCCAUCGCAUUGCGUUGUGCAUUUUUAAUUUGCCUAUGGGC---CAAGGCAGUAAAAAGCACAGCUAAA---GGCGAACAGCUCGCAGCAAAUGGCCAACACAGCCAACAGCCAACA ......((.(((((((((((.((((((...(((((......---..))))).)))))))))))))....---.((((.....)))).)))).((((.......))))...))..... ( -35.80) >DroSim_CAF1 75474 97 + 1 GCCAUCGCAUUGCGUUGUGCAUUUUUAAUUUGCCUACGGGCCGCCAAGGCAGUAAG----UCAGCUGAGUGUGGCCAAAAGCGUAAGGCAAAUGCCCAACG---------------- ((....))....(((((.(((((.......((((((((((((((.....((((...----...))))...)))))).....))).)))))))))).)))))---------------- ( -32.11) >DroEre_CAF1 82796 97 + 1 GCCAUCGCAUUGCGUUGUGCAUUUUUAAUUUGCCUACGGGCCUCCAAGGCAGUAAG----UCAGCUGAGUCUGGCCAAAAGCGUAAGGCAAAUGGCCAACG---------------- (((...((((......)))).......(((((((((((((((.......((((...----...)))).....)))).....))).))))))))))).....---------------- ( -31.20) >DroYak_CAF1 81445 97 + 1 GCCAUCGCAUUGCGUUGUGCAUUUUUAAUUUGCCUACGGGCCGCCAAGGCAGCAAG----UCAGCUGAGUCUGGCCAAAAGCGUAAGGCAAAUGGCCAACG---------------- (((((....((((.((((((...........(((....))).((((...((((...----...))))....)))).....)))))).))))))))).....---------------- ( -33.70) >DroPer_CAF1 78068 111 + 1 GCCAUCGCAUUGCGUUGUGCAUUUUUAAUUUGCCUAUGGGC---CAAGGCAGUAAAAAGCACAGCUAAA---GGCGAACAGCUCGCAGCAAAUGGCCAACACAGCCAACAGCCAACA ......((.(((((((((((.((((((...(((((......---..))))).)))))))))))))....---.((((.....)))).)))).((((.......))))...))..... ( -35.80) >consensus GCCAUCGCAUUGCGUUGUGCAUUUUUAAUUUGCCUACGGGCCGCCAAGGCAGUAAG____UCAGCUGAGU_UGGCCAAAAGCGUAAGGCAAAUGGCCAACG________________ (((((....((((.((((((.........((((((...(.....).))))))...........(((......))).....)))))).)))))))))..................... (-21.53 = -21.92 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:54 2006