
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,449,072 – 7,449,234 |
| Length | 162 |
| Max. P | 0.999433 |

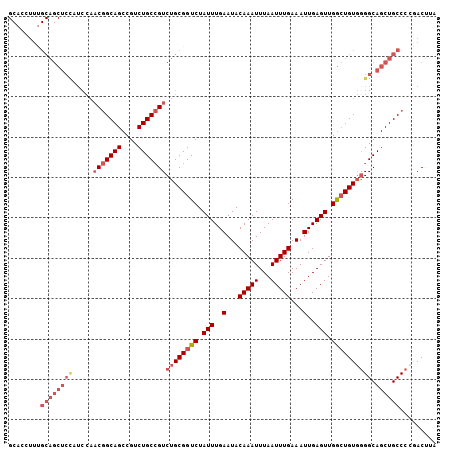
| Location | 7,449,072 – 7,449,178 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 89.87 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -27.40 |
| Energy contribution | -29.32 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7449072 106 - 20766785 GCACCUUUGCAGCUCCAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAUUGAGUUGGCUGUGGGGCAGCUGCCCGAACUUA ........((((((((.....(((((((....)))))))(..(((((.(((..(...(((((...)))))...)..))).)))))..))).))))))......... ( -36.40) >DroSec_CAF1 65506 106 - 1 GCACCUUUGCAGCUCCAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAUUGAGUUGGCUGUGAGGCAGCUGCCCCGACUUA ........((((((((.....(((((((....)))))))..((((((.(((..(...(((((...)))))...)..))).))))))..)).))))))......... ( -33.70) >DroSim_CAF1 57673 106 - 1 GCACCUUUGCAGCUCCAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAUUGAGUUGGCUGUGGGGCAGCUGCCCCGACUUA ........((((((((.....(((((((....)))))))(..(((((.(((..(...(((((...)))))...)..))).)))))..))).))))))......... ( -36.40) >DroEre_CAF1 65643 106 - 1 GCACCUUUGCAGCUCCAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAUUGAGUUGGCUGUGGGGCAGCUGCCCCGACUUA ........((((((((.....(((((((....)))))))(..(((((.(((..(...(((((...)))))...)..))).)))))..))).))))))......... ( -36.40) >DroYak_CAF1 63971 106 - 1 GCACCUUCGCAGCUCUAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAUUGAGUUGGCUGUGGGGCAGCUGCCCCGACUUA ........((((((((.....(((((((....)))))))(..(((((.(((..(...(((((...)))))...)..))).)))))..))).))))))......... ( -34.40) >DroAna_CAF1 62966 85 - 1 ---------------------CCAGCAGCCGUCUGCAGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAUUGAGUUGAAUGUUGGGCAGCUGCCCCAACUUA ---------------------.((((.((((.((((.....)))).)..........((((((((((((......))))))))).)))))).)))).......... ( -22.00) >consensus GCACCUUUGCAGCUCCAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAUUGAGUUGGCUGUGGGGCAGCUGCCCCGACUUA ........((((((((.....(((((((....)))))))((((((((.(((..(...(((((...)))))...)..))).)))))))))).))))))......... (-27.40 = -29.32 + 1.92)


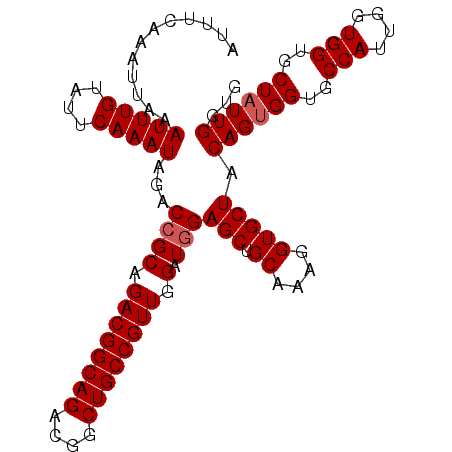
| Location | 7,449,104 – 7,449,209 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 96.95 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -32.86 |
| Energy contribution | -33.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7449104 105 + 20766785 AUUUCAAAUUAAAUUUGUAUUCAAAUAGACCGCAGACGGCAGACGGCUGCCGUUGGAUGGAGCUGCAAAGGUGCUACAGUGGUGCCAUUGGUGGUGCUAUUGUUG ............(((((....)))))...((((.((((((((....)))))))).).)))(((.((....)))))(((((((..(((....)))..))))))).. ( -34.30) >DroSec_CAF1 65538 105 + 1 AUUUCAAAUUAAAUUUGUAUUCAAAUAGACCGCAGACGGCAGACGGCUGCCGUUGGAUGGAGCUGCAAAGGUGCUACAGUGGUGCCAUUGGUGGUGCUAUUGGUG ............(((((....)))))...((((.((((((((....)))))))).).)))(((.((....))))).((((((..(((....)))..))))))... ( -34.10) >DroSim_CAF1 57705 105 + 1 AUUUCAAAUUAAAUUUGUAUUCAAAUAGACCGCAGACGGCAGACGGCUGCCGUUGGAUGGAGCUGCAAAGGUGCUACAGUGGUGCCAUUGGUGGUGCUAUUGGUG ............(((((....)))))...((((.((((((((....)))))))).).)))(((.((....))))).((((((..(((....)))..))))))... ( -34.10) >DroEre_CAF1 65675 104 + 1 AUUUCAAAUUAAAUUUGUAUUCAAAUAGACCGCAGACGGCAGACGGCUGCCGUUGGAUGGAGCUGCAAAGGUGCUACAGUGGUGCCAU-AGUGGUGCUAUUGGUG ............(((((....)))))...((((.((((((((....)))))))).).)))(((.((....))))).((((((..((..-...))..))))))... ( -33.20) >DroYak_CAF1 64003 104 + 1 AUUUCAAAUUAAAUUUGUAUUCAAAUAGACCGCAGACGGCAGACGGCUGCCGUUGGAUAGAGCUGCGAAGGUGCUACAGAGGUGCCAC-AGUGGUGCUAUUGGUG ............(((((....)))))..(((...((((((((....)))))))).(((((.((..(...((..((.....))..))..-.)..)).)))))))). ( -34.10) >consensus AUUUCAAAUUAAAUUUGUAUUCAAAUAGACCGCAGACGGCAGACGGCUGCCGUUGGAUGGAGCUGCAAAGGUGCUACAGUGGUGCCAUUGGUGGUGCUAUUGGUG ............(((((....)))))...((((.((((((((....)))))))).).)))(((.((....))))).((((((..(((....)))..))))))... (-32.86 = -33.26 + 0.40)

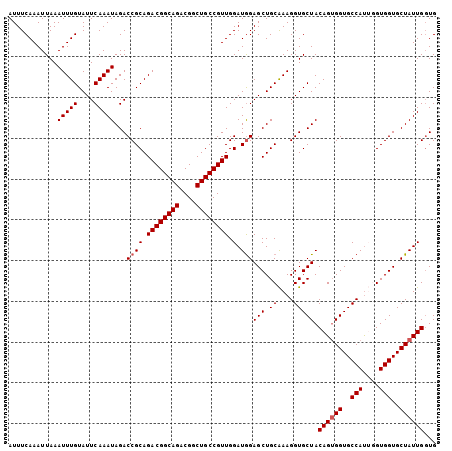
| Location | 7,449,104 – 7,449,209 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 96.95 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -22.76 |
| Energy contribution | -22.96 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7449104 105 - 20766785 CAACAAUAGCACCACCAAUGGCACCACUGUAGCACCUUUGCAGCUCCAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAU ....(((((.(((.....(((.....((((((.....))))))......)))(((((((....)))))))....))))))))......(((((...))))).... ( -23.30) >DroSec_CAF1 65538 105 - 1 CACCAAUAGCACCACCAAUGGCACCACUGUAGCACCUUUGCAGCUCCAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAU ..(.(((((.(((.....(((.....((((((.....))))))......)))(((((((....)))))))....)))))))).)....(((((...))))).... ( -23.90) >DroSim_CAF1 57705 105 - 1 CACCAAUAGCACCACCAAUGGCACCACUGUAGCACCUUUGCAGCUCCAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAU ..(.(((((.(((.....(((.....((((((.....))))))......)))(((((((....)))))))....)))))))).)....(((((...))))).... ( -23.90) >DroEre_CAF1 65675 104 - 1 CACCAAUAGCACCACU-AUGGCACCACUGUAGCACCUUUGCAGCUCCAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAU ..(.(((((.(((.((-((((.....)))))).......((((.........(((((((....))))))))))))))))))).)....(((((...))))).... ( -25.20) >DroYak_CAF1 64003 104 - 1 CACCAAUAGCACCACU-GUGGCACCUCUGUAGCACCUUCGCAGCUCUAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAU ..(.(((((.(((..(-((.(((....))).))).....((((.........(((((((....))))))))))))))))))).)....(((((...))))).... ( -24.70) >consensus CACCAAUAGCACCACCAAUGGCACCACUGUAGCACCUUUGCAGCUCCAUCCAACGGCAGCCGUCUGCCGUCUGCGGUCUAUUUGAAUACAAAUUUAAUUUGAAAU ....(((((.(((.....(((.....((((((.....))))))......)))(((((((....)))))))....))))))))......(((((...))))).... (-22.76 = -22.96 + 0.20)

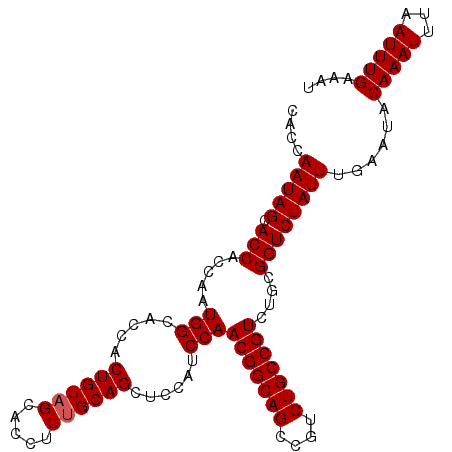
| Location | 7,449,138 – 7,449,234 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -40.48 |
| Energy contribution | -40.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7449138 96 + 20766785 GACGGCAGACGGCUGCCGUUGGAUGGAGCUGCAAAGGUGCUACAGUGGUGCCAUUGGUGGUGCUAUUGUUGGUGCUGCUCAACCACUCAACUGGGC ((((((((....))))))))((...((((......((..(((((((((..(((....)))..)))))).)))..))))))..)).(((....))). ( -41.20) >DroSec_CAF1 65572 96 + 1 GACGGCAGACGGCUGCCGUUGGAUGGAGCUGCAAAGGUGCUACAGUGGUGCCAUUGGUGGUGCUAUUGGUGGUGCUGCUCAACCACUCAACUGGGC ((((((((....))))))))((...((((......((..(((((((((..(((....)))..))))).))))..))))))..)).(((....))). ( -41.20) >DroSim_CAF1 57739 96 + 1 GACGGCAGACGGCUGCCGUUGGAUGGAGCUGCAAAGGUGCUACAGUGGUGCCAUUGGUGGUGCUAUUGGUGGUGCUGCUCAACCACUCAACUGGGC ((((((((....))))))))((...((((......((..(((((((((..(((....)))..))))).))))..))))))..)).(((....))). ( -41.20) >DroEre_CAF1 65709 95 + 1 GACGGCAGACGGCUGCCGUUGGAUGGAGCUGCAAAGGUGCUACAGUGGUGCCAU-AGUGGUGCUAUUGGUGGUGCUGCUCAACCACUCAACUGGGC ((((((((....))))))))((...((((......((..(((((((((..((..-...))..))))).))))..))))))..)).(((....))). ( -40.30) >DroYak_CAF1 64037 95 + 1 GACGGCAGACGGCUGCCGUUGGAUAGAGCUGCGAAGGUGCUACAGAGGUGCCAC-AGUGGUGCUAUUGGUGGUGCUGCUCAACCACUCAACUGGGC ((((((((....)))))))).(((((.((..(...((..((.....))..))..-.)..)).)))))((((((........))))))......... ( -36.70) >consensus GACGGCAGACGGCUGCCGUUGGAUGGAGCUGCAAAGGUGCUACAGUGGUGCCAUUGGUGGUGCUAUUGGUGGUGCUGCUCAACCACUCAACUGGGC ((((((((....))))))))((...((((......((..(((((((((..(((....)))..)))))).)))..))))))..)).(((....))). (-40.48 = -40.68 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:48 2006