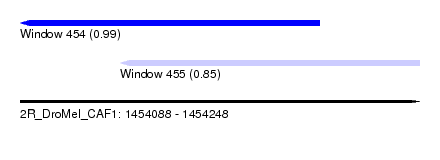
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,454,088 – 1,454,248 |
| Length | 160 |
| Max. P | 0.994362 |

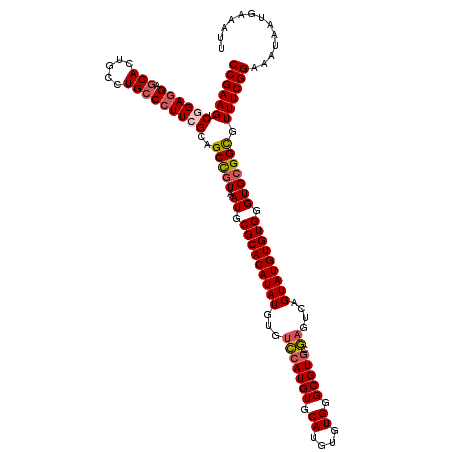
| Location | 1,454,088 – 1,454,208 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -47.68 |
| Consensus MFE | -39.16 |
| Energy contribution | -39.76 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1454088 120 - 20766785 CCGAAGUCGGAGGAGCACUGCCUGCCCUUAGCAGCCGUAAUGCGCACAUAUGUGUCCAUGUGCAUGUGUGGGCGUGCGAGUCAGUAUGUGUGGGUGCGGCGUUUCGGAAAUAAUGAAAUU ((((((.(.((((.(((.....)))))))....((((((.(.(((((((((...(((((((.((....)).))))).))....))))))))).))))))))))))))............. ( -45.30) >DroSec_CAF1 72506 120 - 1 CCGAAGUCGGAGGAGCACUGCCUGCCCUUCGGAGCCGUAAUGCGCACAUAUGUGUCCAUGUACAUGUGUGGGCGUGCGAGUCAGUAUGUGUGGGUGCGGCGUUUCGGAAAUAAUGAAAUU (((((((((((((.(((.....)))))))))).((((((.(.(((((((((...((((((..((....))..)))).))....))))))))).))))))).))))))............. ( -48.60) >DroSim_CAF1 71215 120 - 1 CCGAAGUCGGAGGAGCACUGCCUGCCCUUCGGAGCCGUAAUGCGCACAUAUGUGUCCAUGUACAUGUGUGGGCGUGCGAGUCAGUAUGUGUGGGUGCGGCGUUUCGGAAAUAAUGAAAUU (((((((((((((.(((.....)))))))))).((((((.(.(((((((((...((((((..((....))..)))).))....))))))))).))))))).))))))............. ( -48.60) >DroEre_CAF1 82573 120 - 1 CCGAAGUCGGAGGAGCACUGCCUGCCCUUCGCAGCUGUAAUGCGCACAUAUGUGGCCAUGUGCAUGUGUGGGCGUGCGAGUCAGUAUGUGUGGGUGCGGUGUUUCGGAAAUAAUGAAAUU (((....))).((((((((((..((((..((((((((..((((.(((((((((........))))))))).))))......)))).)))).))))))))))))))............... ( -50.70) >DroYak_CAF1 76709 120 - 1 CCGAAGUCAGAAGAGCACUGCCUGGCCUUCGCAGCUCUGAUGCGCACAUAUGUGUUGAUGUGCAUGUGUGGGCGUGUAAGUCAGUAUGUGUGGGUGCGGUGUUUCGGAAAUAAUGAAAUU (((((.((....))(((((((...((((.((((...(((((((.((((((((..(....)..)))))))).))......)))))..)))).))))))))))))))))............. ( -45.20) >consensus CCGAAGUCGGAGGAGCACUGCCUGCCCUUCGCAGCCGUAAUGCGCACAUAUGUGUCCAUGUGCAUGUGUGGGCGUGCGAGUCAGUAUGUGUGGGUGCGGCGUUUCGGAAAUAAUGAAAUU ((((((.((((((.(((.....)))))))))..(((((.((.(((((((((...(((((((.((....)).))))).))....))))))))).))))))).))))))............. (-39.16 = -39.76 + 0.60)

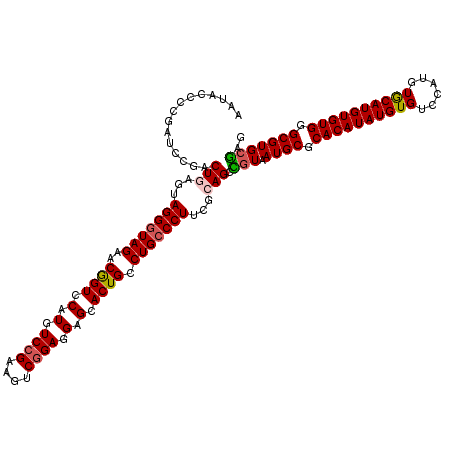
| Location | 1,454,128 – 1,454,248 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -45.88 |
| Consensus MFE | -39.40 |
| Energy contribution | -39.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1454128 120 - 20766785 AAUACCCCGAUCCUACUGAGUAGGGUAGAACGGUCCAUGUCCGAAGUCGGAGGAGCACUGCCUGCCCUUAGCAGCCGUAAUGCGCACAUAUGUGUCCAUGUGCAUGUGUGGGCGUGCGAG ...............(((...(((((((..((((.(.(.((((....)))).).).)))).)))))))...))).((((.(((.((((((((..(....)..)))))))).))))))).. ( -46.00) >DroSec_CAF1 72546 120 - 1 AAUACCCCGAUCCGACUGAGUAGGGUAGAACGGUCCAUGUCCGAAGUCGGAGGAGCACUGCCUGCCCUUCGGAGCCGUAAUGCGCACAUAUGUGUCCAUGUACAUGUGUGGGCGUGCGAG ..........(((((......(((((((..((((.(.(.((((....)))).).).)))).))))))))))))..((((.(((.((((((((((......)))))))))).))))))).. ( -47.80) >DroSim_CAF1 71255 120 - 1 AAUACCCCGAUCCGACUGAGUAGGGUAGAACGGUCCAUGUCCGAAGUCGGAGGAGCACUGCCUGCCCUUCGGAGCCGUAAUGCGCACAUAUGUGUCCAUGUACAUGUGUGGGCGUGCGAG ..........(((((......(((((((..((((.(.(.((((....)))).).).)))).))))))))))))..((((.(((.((((((((((......)))))))))).))))))).. ( -47.80) >DroEre_CAF1 82613 120 - 1 AAUACCCCAAACCGGCUGAGUAGGGUAGAACGGUCCAUGUCCGAAGUCGGAGGAGCACUGCCUGCCCUUCGCAGCUGUAAUGCGCACAUAUGUGGCCAUGUGCAUGUGUGGGCGUGCGAG ............((((((...(((((((..((((.(.(.((((....)))).).).)))).)))))))...))))))..((((.(((((((((........))))))))).))))..... ( -49.60) >DroYak_CAF1 76749 115 - 1 AAUACCCC-----GACUGUGUAGGGUAGAACAGUCCAUGUCCGAAGUCAGAAGAGCACUGCCUGGCCUUCGCAGCUCUGAUGCGCACAUAUGUGUUGAUGUGCAUGUGUGGGCGUGUAAG ..(((...-----(((((((.(((.(((..((((.(...((.(....).))...).)))).))).))).))))).))..((((.((((((((..(....)..)))))))).))))))).. ( -38.20) >consensus AAUACCCCGAUCCGACUGAGUAGGGUAGAACGGUCCAUGUCCGAAGUCGGAGGAGCACUGCCUGCCCUUCGCAGCCGUAAUGCGCACAUAUGUGUCCAUGUGCAUGUGUGGGCGUGCGAG ...............(((...(((((((..((((.(.(.((((....)))).).).)))).)))))))...))).(((.((((.((((((((((......)))))))))).))))))).. (-39.40 = -39.60 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:11 2006