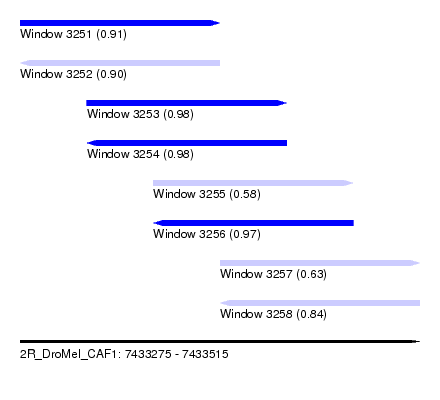
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,433,275 – 7,433,515 |
| Length | 240 |
| Max. P | 0.980976 |

| Location | 7,433,275 – 7,433,395 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.79 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -32.38 |
| Energy contribution | -32.98 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7433275 120 + 20766785 CUGGCCUCCAAGUGGGCCCAUUAAUCAGUGGCCCCCACAUACAAUACAUACGGUGCAUAUAUCACAUGGGCAGGCGCCCACGCUUUUGGGAUCCCACUGGGGGAUGCACUCUAAUGAUUU ..(((((......)))))(((((...((((.((((((.............(((((.......))).(((((....)))))))....(((....))).))))))...)))).))))).... ( -38.10) >DroSec_CAF1 49625 110 + 1 CUGGCCUCCAAGUGGGCCCAUUAAUCAGAGGCCCCCACA---------UAGGGGACAUAUAUCACAUGGGCAGGCGCCCACGCUU-UGGGAUCCCACUGGGGGAUGCACUCUAAUGAUUU ....((((((.(((((((((........((((((((...---------..))))............(((((....))))).))))-))))..)))))))))))................. ( -41.10) >DroSim_CAF1 41206 110 + 1 CUGGCCUCCAAGUGGGCCCAUUAAUCAGAGGCCCCCACA---------UAGGGGGCAUAUAUCACAUGGGCAGGCGCCCACGCUU-UGGGAUCCAACUGGGGGAUGCACUCUAAUGAUUU ....((((((..((((((((.......((.((((((...---------..)))))).....))...(((((....))))).....-)))).))))..))))))................. ( -44.80) >DroEre_CAF1 49339 105 + 1 CUGGCCUCCAAGUGGGCCCAUUAAUCAGAGGCCCCCACA---------UACGGGG-AUAUAUC--AUGGGCAGGCGCCCACGCUU-UGGGAUACCAUU--GGGAUGCACUCUAAUGAUUU ...((.(((.(((((.((((........((((((((...---------...))))-.......--.(((((....))))).))))-))))...)))))--.))).))............. ( -36.80) >DroYak_CAF1 47803 106 + 1 CUGGCCUCCAAGUGGGCCCAUUAAUCAGAGGCCCCCACA---------UACGGGGCAUAUAUC--AUGGGUUGGCGCCCACGCUU-GGGGAUCCCAUG--GGGAUGCACUCUAAUGAUUU ...(((((((((((((((((....(((((.(((((....---------...))))).....))--.)))..))).))))..))))-))))(((((...--)))))))............. ( -41.30) >consensus CUGGCCUCCAAGUGGGCCCAUUAAUCAGAGGCCCCCACA_________UACGGGGCAUAUAUCACAUGGGCAGGCGCCCACGCUU_UGGGAUCCCACUGGGGGAUGCACUCUAAUGAUUU ...((.(((.(((((((((((.........(((((................))))).........)))))).....(((........)))...)))))...))).))............. (-32.38 = -32.98 + 0.60)

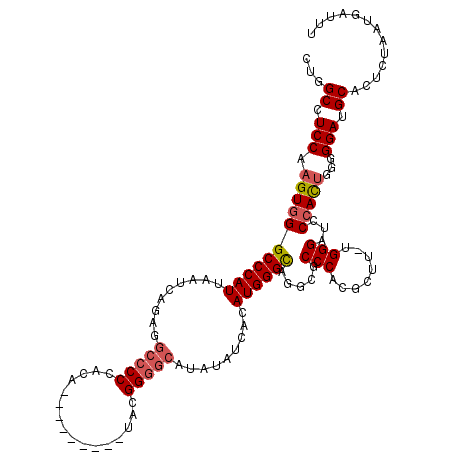
| Location | 7,433,275 – 7,433,395 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.79 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -32.16 |
| Energy contribution | -34.60 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7433275 120 - 20766785 AAAUCAUUAGAGUGCAUCCCCCAGUGGGAUCCCAAAAGCGUGGGCGCCUGCCCAUGUGAUAUAUGCACCGUAUGUAUUGUAUGUGGGGGCCACUGAUUAAUGGGCCCACUUGGAGGCCAG ....((((((((((...(((((((..((..((((......))))..))..).((((..((((((((...))))))))..)))))))))).))))..))))))((((........)))).. ( -48.30) >DroSec_CAF1 49625 110 - 1 AAAUCAUUAGAGUGCAUCCCCCAGUGGGAUCCCA-AAGCGUGGGCGCCUGCCCAUGUGAUAUAUGUCCCCUA---------UGUGGGGGCCUCUGAUUAAUGGGCCCACUUGGAGGCCAG .....(((((((.((..((((((..(((((....-..((((((((....)))))))).......)))))...---------)).))))))))))))).....((((........)))).. ( -46.42) >DroSim_CAF1 41206 110 - 1 AAAUCAUUAGAGUGCAUCCCCCAGUUGGAUCCCA-AAGCGUGGGCGCCUGCCCAUGUGAUAUAUGCCCCCUA---------UGUGGGGGCCUCUGAUUAAUGGGCCCACUUGGAGGCCAG ..................((((((.(((..((((-..((((((((....)))))))).......((((((..---------...))))))..........)))).))).)))).)).... ( -42.90) >DroEre_CAF1 49339 105 - 1 AAAUCAUUAGAGUGCAUCCC--AAUGGUAUCCCA-AAGCGUGGGCGCCUGCCCAU--GAUAUAU-CCCCGUA---------UGUGGGGGCCUCUGAUUAAUGGGCCCACUUGGAGGCCAG .....(((((((.((.((((--(((((.......-...(((((((....))))))--)......-..)))).---------..)))))))))))))).....((((........)))).. ( -37.67) >DroYak_CAF1 47803 106 - 1 AAAUCAUUAGAGUGCAUCCC--CAUGGGAUCCCC-AAGCGUGGGCGCCAACCCAU--GAUAUAUGCCCCGUA---------UGUGGGGGCCUCUGAUUAAUGGGCCCACUUGGAGGCCAG .....(((((((.(((((((--...)))))((((-...((((((......)))))--)(((((((...))))---------)))))))))))))))).....((((........)))).. ( -40.70) >consensus AAAUCAUUAGAGUGCAUCCCCCAGUGGGAUCCCA_AAGCGUGGGCGCCUGCCCAUGUGAUAUAUGCCCCGUA_________UGUGGGGGCCUCUGAUUAAUGGGCCCACUUGGAGGCCAG .....(((((((.(((((((.....))))).......((((((((....))))))))........(((((((((.....)))))))))))))))))).....((((........)))).. (-32.16 = -34.60 + 2.44)

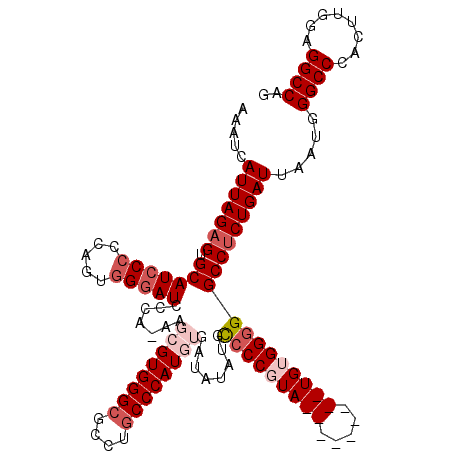
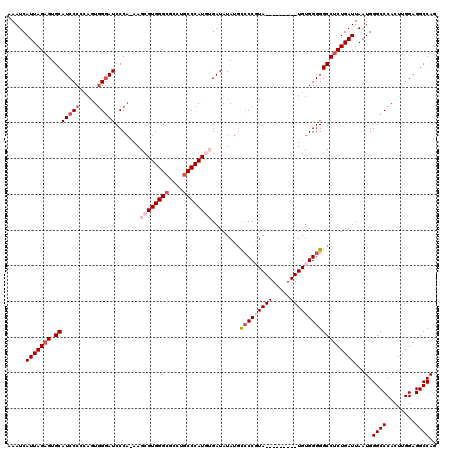
| Location | 7,433,315 – 7,433,435 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.97 |
| Mean single sequence MFE | -37.96 |
| Consensus MFE | -30.50 |
| Energy contribution | -31.14 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7433315 120 + 20766785 ACAAUACAUACGGUGCAUAUAUCACAUGGGCAGGCGCCCACGCUUUUGGGAUCCCACUGGGGGAUGCACUCUAAUGAUUUUAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGA ........((.(((((((........(((((....)))))(.((..(((....)))..)).).))))))).))(((((((.(((((.......)))))))))))).((((.....)))). ( -40.80) >DroSec_CAF1 49664 111 + 1 --------UAGGGGACAUAUAUCACAUGGGCAGGCGCCCACGCUU-UGGGAUCCCACUGGGGGAUGCACUCUAAUGAUUUUAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGA --------(((((.............(((((....))))).....-..(.(((((.....))))).).)))))(((((((.(((((.......)))))))))))).((((.....)))). ( -38.00) >DroSim_CAF1 41245 111 + 1 --------UAGGGGGCAUAUAUCACAUGGGCAGGCGCCCACGCUU-UGGGAUCCAACUGGGGGAUGCACUCUAAUGAUUUUAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGA --------(((((.((((........(((((....)))))..(((-..(.......)..))).)))).)))))(((((((.(((((.......)))))))))))).((((.....)))). ( -38.50) >DroEre_CAF1 49378 106 + 1 --------UACGGGG-AUAUAUC--AUGGGCAGGCGCCCACGCUU-UGGGAUACCAUU--GGGAUGCACUCUAAUGAUUUUAUGGCAAUGCUCGCCAUAAAUCAUACCAGUCGAACUGGA --------...(((.-.(((...--.(((((....)))))(.(..-(((....)))..--).))))..)))..(((((((.(((((.......)))))))))))).((((.....)))). ( -34.40) >DroYak_CAF1 47842 107 + 1 --------UACGGGGCAUAUAUC--AUGGGUUGGCGCCCACGCUU-GGGGAUCCCAUG--GGGAUGCACUCUAAUGAUUUUAUGGCAAUGUUCGCCAUAAAUCAUCGCAGUCGAACUGGA --------...(((((((...((--(((((....(.((((....)-))))..))))))--)..)))).)))..(((((((.(((((.......))))))))))))..(((.....))).. ( -38.10) >consensus ________UACGGGGCAUAUAUCACAUGGGCAGGCGCCCACGCUU_UGGGAUCCCACUGGGGGAUGCACUCUAAUGAUUUUAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGA ...........(((............(((((....)))))........(.(((((.....))))).).)))..(((((((.(((((.......)))))))))))).((((.....)))). (-30.50 = -31.14 + 0.64)

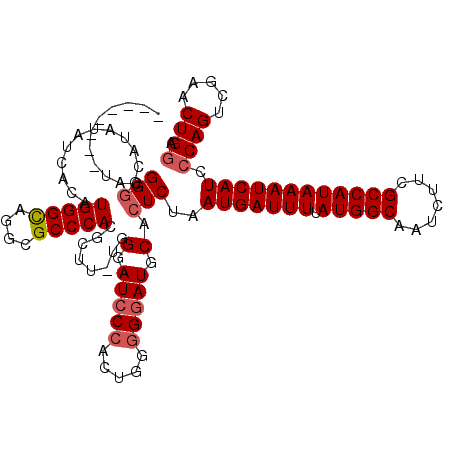
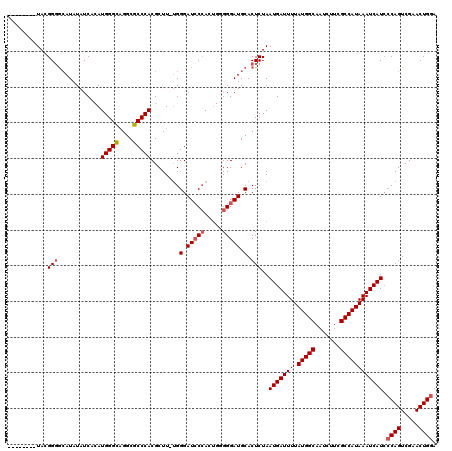
| Location | 7,433,315 – 7,433,435 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.97 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -29.70 |
| Energy contribution | -31.50 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.980976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7433315 120 - 20766785 UCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUAAAAUCAUUAGAGUGCAUCCCCCAGUGGGAUCCCAAAAGCGUGGGCGCCUGCCCAUGUGAUAUAUGCACCGUAUGUAUUGU ....((((.((((((((((((((((((((...))))))).)))))))..((.....)).)))))).)))).......((((((((....))))))))(((((((((...))))))))).. ( -45.00) >DroSec_CAF1 49664 111 - 1 UCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUAAAAUCAUUAGAGUGCAUCCCCCAGUGGGAUCCCA-AAGCGUGGGCGCCUGCCCAUGUGAUAUAUGUCCCCUA-------- .........((((((((((((((((((((...))))))).)))))))..((.....)).))))))(((((....-..((((((((....)))))))).......)))))...-------- ( -42.22) >DroSim_CAF1 41245 111 - 1 UCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUAAAAUCAUUAGAGUGCAUCCCCCAGUUGGAUCCCA-AAGCGUGGGCGCCUGCCCAUGUGAUAUAUGCCCCCUA-------- ....(((((((((((((((((((((((((...))))))).)))))))..((.....)).)))))))))))....-..((((((((....))))))))...............-------- ( -41.30) >DroEre_CAF1 49378 106 - 1 UCCAGUUCGACUGGUAUGAUUUAUGGCGAGCAUUGCCAUAAAAUCAUUAGAGUGCAUCCC--AAUGGUAUCCCA-AAGCGUGGGCGCCUGCCCAU--GAUAUAU-CCCCGUA-------- .((((.....)))).((((((((((((((...))))))).))))))).............--.((((.......-...(((((((....))))))--)......-..)))).-------- ( -30.67) >DroYak_CAF1 47842 107 - 1 UCCAGUUCGACUGCGAUGAUUUAUGGCGAACAUUGCCAUAAAAUCAUUAGAGUGCAUCCC--CAUGGGAUCCCC-AAGCGUGGGCGCCAACCCAU--GAUAUAUGCCCCGUA-------- .........(((..(((((((((((((((...))))))).))))))))..)))((((...--((((((..((((-(....)))).)....)))))--)....))))......-------- ( -32.30) >consensus UCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUAAAAUCAUUAGAGUGCAUCCCCCAGUGGGAUCCCA_AAGCGUGGGCGCCUGCCCAUGUGAUAUAUGCCCCGUA________ .((((.....))))(((((((((((((((...))))))).))))))))...(.(.(((((.....))))).))....((((((((....))))))))....................... (-29.70 = -31.50 + 1.80)

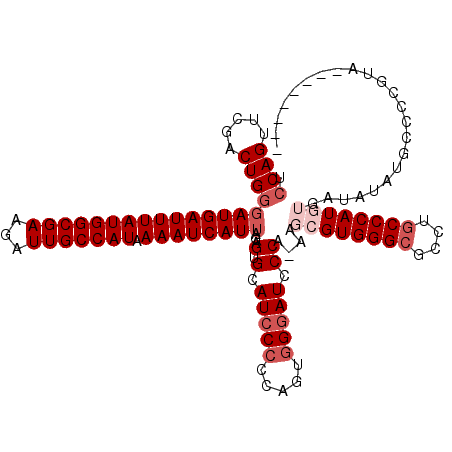
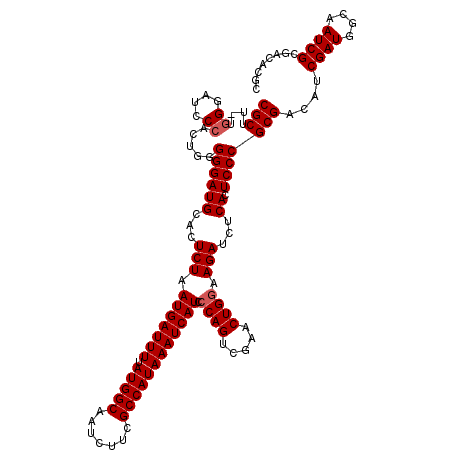
| Location | 7,433,355 – 7,433,475 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.80 |
| Mean single sequence MFE | -35.56 |
| Consensus MFE | -30.22 |
| Energy contribution | -30.62 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7433355 120 + 20766785 CGCUUUUGGGAUCCCACUGGGGGAUGCACUCUAAUGAUUUUAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGAAGAUCUCACUCCCGCGACAUCGAUGGCAAUCGCGACACGC (((...(((....)))..(((((.((...(((.(((((((.(((((.......)))))))))))).((((.....)))).)))...))))))))))....((((....))))........ ( -35.60) >DroSec_CAF1 49696 119 + 1 CGCUU-UGGGAUCCCACUGGGGGAUGCACUCUAAUGAUUUUAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGAAGAUCUCACUCCCGCGACAUCGAUGGCAAUCGCGACACGC (((..-(((....)))..(((((.((...(((.(((((((.(((((.......)))))))))))).((((.....)))).)))...))))))))))....((((....))))........ ( -35.60) >DroSim_CAF1 41277 119 + 1 CGCUU-UGGGAUCCAACUGGGGGAUGCACUCUAAUGAUUUUAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGAAGAUCUCACUCCCGCGACAUCGAUGGCAAUCGCGACACGC (((..-(((((((....(((((......)))))(((((((.(((((.......)))))))))))).((((.....))))..)))))))...(((((....)).))).....)))...... ( -35.30) >DroEre_CAF1 49407 117 + 1 CGCUU-UGGGAUACCAUU--GGGAUGCACUCUAAUGAUUUUAUGGCAAUGCUCGCCAUAAAUCAUACCAGUCGAACUGGAAGAUCUCACUCCCGCGACUUCGAUGGCAAUCGCGACACGC (((..-(((....)))..--((((((...(((.(((((((.(((((.......)))))))))))).((((.....)))).)))...)).)))))))....((((....))))........ ( -33.70) >DroYak_CAF1 47872 117 + 1 CGCUU-GGGGAUCCCAUG--GGGAUGCACUCUAAUGAUUUUAUGGCAAUGUUCGCCAUAAAUCAUCGCAGUCGAACUGGAAGAUCUCACUCCCGCGACUUCGAUGGCAAUCGCGACACGC (((((-(((((((((...--)))))...)))))).(((((((((((.......))))))).((((((.(((((....(((.........)))..))))).)))))).)))))))...... ( -37.60) >consensus CGCUU_UGGGAUCCCACUGGGGGAUGCACUCUAAUGAUUUUAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGAAGAUCUCACUCCCGCGACAUCGAUGGCAAUCGCGACACGC (((....((....)).....((((((...(((.(((((((.(((((.......)))))))))))).((((.....)))).)))...)).)))))))....((((....))))........ (-30.22 = -30.62 + 0.40)


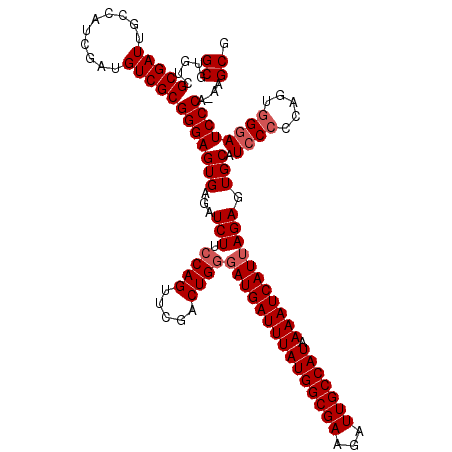
| Location | 7,433,355 – 7,433,475 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.80 |
| Mean single sequence MFE | -41.36 |
| Consensus MFE | -36.28 |
| Energy contribution | -37.08 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7433355 120 - 20766785 GCGUGUCGCGAUUGCCAUCGAUGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUAAAAUCAUUAGAGUGCAUCCCCCAGUGGGAUCCCAAAAGCG (((((.((((((....)))).)).)))(((((((...(((.((((.....))))(((((((((((((((...))))))).))))))))))).))).((((.....))))))))....)). ( -42.50) >DroSec_CAF1 49696 119 - 1 GCGUGUCGCGAUUGCCAUCGAUGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUAAAAUCAUUAGAGUGCAUCCCCCAGUGGGAUCCCA-AAGCG (((((.((((((....)))).)).)))(((((((...(((.((((.....))))(((((((((((((((...))))))).))))))))))).))).((((.....)))))))).-..)). ( -42.50) >DroSim_CAF1 41277 119 - 1 GCGUGUCGCGAUUGCCAUCGAUGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUAAAAUCAUUAGAGUGCAUCCCCCAGUUGGAUCCCA-AAGCG (((((.((((((....)))).)).)))(((((......))))).(((((((((((((((((((((((((...))))))).)))))))..((.....)).)))))))))))....-..)). ( -43.30) >DroEre_CAF1 49407 117 - 1 GCGUGUCGCGAUUGCCAUCGAAGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGGUAUGAUUUAUGGCGAGCAUUGCCAUAAAAUCAUUAGAGUGCAUCCC--AAUGGUAUCCCA-AAGCG ((.....((((((........))))))((((.((...(((.((((.....)))).((((((((((((((...))))))).))))))).)))...))))))--..(((....)))-..)). ( -37.70) >DroYak_CAF1 47872 117 - 1 GCGUGUCGCGAUUGCCAUCGAAGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGCGAUGAUUUAUGGCGAACAUUGCCAUAAAAUCAUUAGAGUGCAUCCC--CAUGGGAUCCCC-AAGCG (((.(((((((((........))))))(((((......))))).....))))))(((((((((((((((...))))))).)))))))).(..((.(((((--...)))))...)-)..). ( -40.80) >consensus GCGUGUCGCGAUUGCCAUCGAUGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUAAAAUCAUUAGAGUGCAUCCCCCAGUGGGAUCCCA_AAGCG ((.....(((((..........)))))(((((((...(((.((((.....))))(((((((((((((((...))))))).))))))))))).))).((((.....))))))))....)). (-36.28 = -37.08 + 0.80)


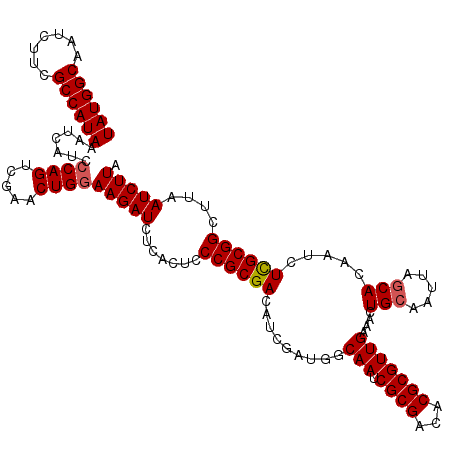
| Location | 7,433,395 – 7,433,515 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -27.72 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7433395 120 + 20766785 UAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGAAGAUCUCACUCCCGCGACAUCGAUGGCAAUCGCGACACGCGUUGAAACUGCAAUUAGCACAAUCUUGCGGCUUAAUCUUA ((((((.......)))))).......((((.....))))(((((.......((((((.........(((.((((...)))))))....(((.....))).....))))))....))))). ( -28.60) >DroSec_CAF1 49735 120 + 1 UAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGAAGAUCUCACUCCCGCGACAUCGAUGGCAAUCGCGACACGCGUUGAAACUGCAAUUAGCACAAUCUCGCGGCUUAAUCUUA ((((((.......)))))).......((((.....))))(((((.......((((((.........(((.((((...)))))))....(((.....))).....))))))....))))). ( -30.70) >DroSim_CAF1 41316 120 + 1 UAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGAAGAUCUCACUCCCGCGACAUCGAUGGCAAUCGCGACACGCGUUGAAACUGCAAUUAGCACAAUCUUGCGGCUUAAUCUUA ((((((.......)))))).......((((.....))))(((((.......((((((.........(((.((((...)))))))....(((.....))).....))))))....))))). ( -28.60) >DroEre_CAF1 49444 120 + 1 UAUGGCAAUGCUCGCCAUAAAUCAUACCAGUCGAACUGGAAGAUCUCACUCCCGCGACUUCGAUGGCAAUCGCGACACGCGUUGAAGCUGCAAUUAACACAAUCUCGCGGCCUAAUCUUA ((((((.......)))))).......((((.....))))(((((.......((((((....(((....)))(((...)))(((((........)))))......))))))....))))). ( -29.30) >DroYak_CAF1 47909 120 + 1 UAUGGCAAUGUUCGCCAUAAAUCAUCGCAGUCGAACUGGAAGAUCUCACUCCCGCGACUUCGAUGGCAAUCGCGACACGCGUUGAAACUGCAAUUAGCACAAUCUCGCGGCUUAAUCUUA ((((((.......))))))...(((((.(((((....(((.........)))..))))).)))))((....))....((((..((...(((.....)))...)).))))........... ( -32.60) >consensus UAUGGCAAUCUUCGCCAUAAAUCAUCCCAGUCGAACUGGAAGAUCUCACUCCCGCGACAUCGAUGGCAAUCGCGACACGCGUUGAAACUGCAAUUAGCACAAUCUCGCGGCUUAAUCUUA ((((((.......)))))).......((((.....))))(((((.......((((((.........(((.((((...)))))))....(((.....))).....))))))....))))). (-27.72 = -27.88 + 0.16)

| Location | 7,433,395 – 7,433,515 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -34.78 |
| Energy contribution | -35.82 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7433395 120 - 20766785 UAAGAUUAAGCCGCAAGAUUGUGCUAAUUGCAGUUUCAACGCGUGUCGCGAUUGCCAUCGAUGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUA ...(((((.((.(((....))))))))))(((((((...((((...)))).(((((((.(((.((((....)))).)))((((((.....))))))......)))))))))))))).... ( -38.30) >DroSec_CAF1 49735 120 - 1 UAAGAUUAAGCCGCGAGAUUGUGCUAAUUGCAGUUUCAACGCGUGUCGCGAUUGCCAUCGAUGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUA ..........((((((.((((((.(((((((................))))))).)).)))).)))))).....(((((((((((.....)))))).)))))(((((((...))))))). ( -40.29) >DroSim_CAF1 41316 120 - 1 UAAGAUUAAGCCGCAAGAUUGUGCUAAUUGCAGUUUCAACGCGUGUCGCGAUUGCCAUCGAUGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUA ...(((((.((.(((....))))))))))(((((((...((((...)))).(((((((.(((.((((....)))).)))((((((.....))))))......)))))))))))))).... ( -38.30) >DroEre_CAF1 49444 120 - 1 UAAGAUUAGGCCGCGAGAUUGUGUUAAUUGCAGCUUCAACGCGUGUCGCGAUUGCCAUCGAAGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGGUAUGAUUUAUGGCGAGCAUUGCCAUA ........((((((((.((.(((((.(........).))))))).))))).(((((((.....((((....))))((((..((((.....))))...)))).))))))).....)))... ( -37.90) >DroYak_CAF1 47909 120 - 1 UAAGAUUAAGCCGCGAGAUUGUGCUAAUUGCAGUUUCAACGCGUGUCGCGAUUGCCAUCGAAGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGCGAUGAUUUAUGGCGAACAUUGCCAUA ..((((((......((((((((.......))))))))..((((.(((((((((........))))))(((((......))))).....))))))).))))))(((((((...))))))). ( -39.70) >consensus UAAGAUUAAGCCGCGAGAUUGUGCUAAUUGCAGUUUCAACGCGUGUCGCGAUUGCCAUCGAUGUCGCGGGAGUGAGAUCUUCCAGUUCGACUGGGAUGAUUUAUGGCGAAGAUUGCCAUA ..........((((((.((((((.(((((((.((......)).....))))))).)).)))).)))))).....(((((((((((.....)))))).)))))(((((((...))))))). (-34.78 = -35.82 + 1.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:42 2006