
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,420,586 – 7,420,696 |
| Length | 110 |
| Max. P | 0.783628 |

| Location | 7,420,586 – 7,420,696 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.84 |
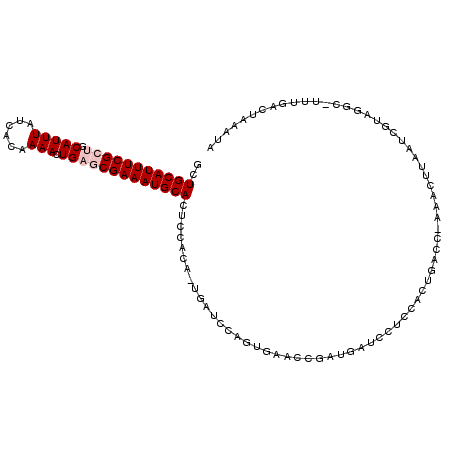
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -12.50 |
| Energy contribution | -13.17 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783628 |
| Prediction | RNA |
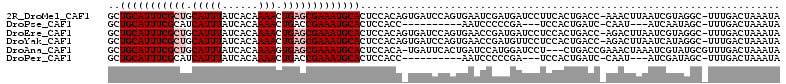
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7420586 110 + 20766785 GCUGCAUUUCGCUGCAUUUAUCACAAAACUGAGCGAAAUGCACUCCACAGUGAUCCAGUGAAUCGAUGAUCCUUCACUGACC-AAACUUAAUCGUAGGC-UUUGACUAAAUA ..(((((((((((.(((((......))).)))))))))))))......(((....(((((((..........)))))))..(-(((((((....)))).-)))))))..... ( -26.00) >DroPse_CAF1 36630 94 + 1 GCUGCAUUUCGCAUCAUUUAUCACAAAACUGACCGAAAUGCACUCCACC----------AAUCCCCCGA---UCCACUGAUC-CAAU---AUCAAUAGC-UUUGACUAAAUA (((((((((((..((((((......))).))).))))))))........----------........((---((....))))-....---......)))-............ ( -15.30) >DroEre_CAF1 36608 110 + 1 GCUGCAUUUCGCUGCAUUUAUCACAAAACUGAGCGAAAUGCACUCCACAGUGAUCCAGUGAACCGAUGAUCCUCCACUGACC-AGACUUAAUCGUAGGC-UUUGACUAAAUA ..(((((((((((.(((((......))).)))))))))))))......(((....(((((..............)))))..(-(((((((....)))).-)))))))..... ( -23.64) >DroYak_CAF1 35113 110 + 1 GCUGCAUUUCGCUGCAUUUAUCACAAAACUGAGCGAAAUGCACUCCACAGUGAUCCAGUGAACCGAUGUUCCUCCACUGACC-AGACUUAAUCAUAGGC-UUUGACUAAAUA ..(((((((((((.(((((......))).))))))))))))).....(((((....((.((((....)))))).)))))..(-(((((((....)))).-))))........ ( -26.70) >DroAna_CAF1 35050 108 + 1 GCUGCAUUUCGCUGCAUUUAUCACAAAAGUGAGCGAAAUGCACUCCACA-UGAUUCACUGAUCCAUGGAUCCU---CUGACCGAAACUAAAUCGUAUGCGUUUGACUAAAUA (((((((((((((.(((((.......))))))))))))))))......(-(((((....((((....)))).(---(.....)).....))))))..))............. ( -26.40) >DroPer_CAF1 36592 94 + 1 GCUGCAUUUCGCAUCAUUUAUCACAAAACUGACCGAAAUGCACUCCACC----------AAUCCCCCGA---UCCACUGAUC-CAAU---AUCGAUAGC-UUUGACUAAAUA (((((((((((..((((((......))).))).))))))))........----------........((---((....))))-....---......)))-............ ( -15.30) >consensus GCUGCAUUUCGCUGCAUUUAUCACAAAACUGAGCGAAAUGCACUCCACA_UGAUCCAGUGAACCGAUGAUCCUCCACUGACC_AAACUUAAUCGUAGGC_UUUGACUAAAUA ..(((((((((((.(((((......))).)))))))))))))...................................................................... (-12.50 = -13.17 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:32 2006