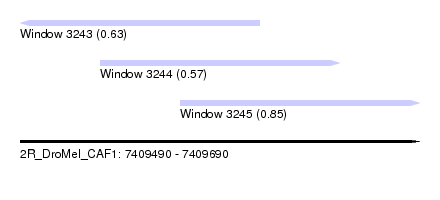
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,409,490 – 7,409,690 |
| Length | 200 |
| Max. P | 0.854667 |

| Location | 7,409,490 – 7,409,610 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -32.86 |
| Energy contribution | -32.58 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
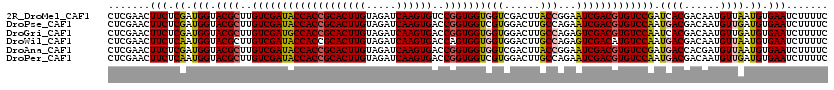
>2R_DroMel_CAF1 7409490 120 - 20766785 CUCGAACUUCUCGAUGGUACGCUUGUCGAUACCACCGCACUUGUAGAUCAAGUGUCCGGUGGUGGUCGACUUACCGGAAUCGACGUGUCCGAUCACGACAAUGUUAAUGUGAAUCUUUUC .(((.((...(((((((((.....(((((((((((((((((((.....))))))..))))))).)))))).))))...)))))...)).)))(((((..........)))))........ ( -39.50) >DroPse_CAF1 25290 120 - 1 CUCGAACUUCUCGAUGGUACGCUUGUCGAUACCACCGCACUUGUAGAUCAAGUGACCGGUGGUCGUGGACUUGCCAGAAUCGACGUGUCCAAUGACGACAAUGUUGAUGUGAAUCUUUUC ............(((..(((((..(((.(((((((((((((((.....))))))..))))))).)).)))..))....(((((((((((.......))).))))))))))).)))..... ( -40.10) >DroGri_CAF1 23959 120 - 1 CUCGAACUUCUCGAUGGUACGCUUGUCGAUGCCACCGCACUUGUAGAUCAAGUGACCGGUGGUGGUGGACUUGCCAGAGUCGACGUGUCCAAUCACGACAAUGUUGAUGUGAAUCUUUUC .((((.....))))(((.((((..(((((((((((((((((((.....))))))..)))))))(((......)))...))))))))))))).(((((.((....)).)))))........ ( -42.80) >DroWil_CAF1 27469 120 - 1 CUCGAACUUCUCAAUGGUACGCUUGUCGAUACCACCGCACUUGUAGAUCAAGUGACCAGUGGUGGUGGACUUGCCAGAGUCGACAUGUCCAAUGACGACAAUGUUAAUGUGAAUCUUUUC ....(((.(((((.(((.(((..((((..((((((((((((((.....))))).....)))))))))(((((....))))))))))))))).))).))....)))............... ( -34.00) >DroAna_CAF1 24480 120 - 1 CUCGAACUUCUCGAUGGUACGCUUGUCGAUACCACCGCACUUGUAGAUCAAGUGACCGGUGGUGGUCGACUUACCGGAAUCGACGUGUCCGAUGACCACGAUGUUAAUGUGAAUCUUUUC .(((.((...(((((((((.....(((((((((((((((((((.....))))))..))))))).)))))).))))...)))))...)).)))....((((.......))))......... ( -38.50) >DroPer_CAF1 25242 120 - 1 CUCGAACUUCUCAAUGGUACGCUUGUCGAUACCACCGCACUUGUAGAUCAAGUGACCGGUGGUCGUGGACUUGCCAGAAUCGACGUGUCCAAUGACGACAAUGUUGAUGUGAAUCUUUUC .((((...(((((.(((.((((..(((((((((((((((((((.....))))))..)))))))..(((.....)))..))))))))))))).))).)).....))))............. ( -38.30) >consensus CUCGAACUUCUCGAUGGUACGCUUGUCGAUACCACCGCACUUGUAGAUCAAGUGACCGGUGGUGGUGGACUUGCCAGAAUCGACGUGUCCAAUGACGACAAUGUUAAUGUGAAUCUUUUC .......(((.((.(((.((((..(((((((((((((((((((.....))))))..)))))))(((......)))...))))))))))))).((((......)))).)).)))....... (-32.86 = -32.58 + -0.28)
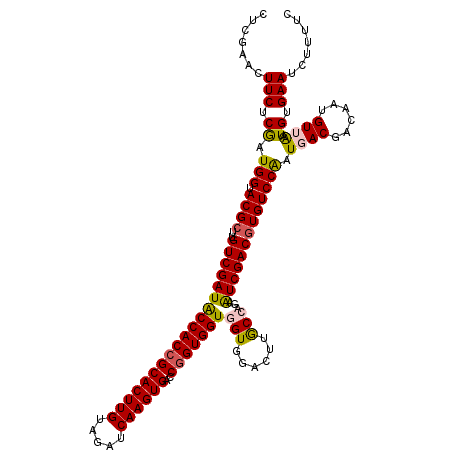
| Location | 7,409,530 – 7,409,650 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -41.63 |
| Consensus MFE | -34.62 |
| Energy contribution | -34.23 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7409530 120 + 20766785 AUUCCGGUAAGUCGACCACCACCGGACACUUGAUCUACAAGUGCGGUGGUAUCGACAAGCGUACCAUCGAGAAGUUCGAGAAGGAGGCCCAGGAGAUGGGAAAGGGAUCCUUCAAGUACG ..........(((((..(((((((..((((((.....))))))))))))).)))))...(((((..(((((...)))))((((((..((((.....)))).......))))))..))))) ( -46.40) >DroVir_CAF1 23871 120 + 1 ACUCUGGCAAGUCCACCACCACCGGUCACUUGAUCUACAAGUGCGGUGGUAUCGACAAGCGUACCAUUGAGAAGUUCGAGAAGGAGGCUCAGGAAAUGGGCAAGGGCUCGUUCAAGUACG (((..(((.(((((.(((((((((..((((((.....))))))))))))).((((....((......))......))))...))..(((((.....)))))..))))).)))..)))... ( -39.50) >DroGri_CAF1 23999 120 + 1 ACUCUGGCAAGUCCACCACCACCGGUCACUUGAUCUACAAGUGCGGUGGCAUCGACAAGCGUACCAUCGAGAAGUUCGAGAAGGAAGCUCAGGAAAUGGGCAAGGGCUCGUUCAAGUACG (((..(((.(((((.((.((((((..((((((.....))))))))))))..((((....((......))......))))...))..(((((.....)))))..))))).)))..)))... ( -39.30) >DroWil_CAF1 27509 120 + 1 ACUCUGGCAAGUCCACCACCACUGGUCACUUGAUCUACAAGUGCGGUGGUAUCGACAAGCGUACCAUUGAGAAGUUCGAGAAGGAAGCUCAGGAAAUGGGCAAGGGUUCCUUCAAGUACG ......((..(((....(((((((..((((((.....)))))))))))))...)))..))((((..(((((...)))))((((((((((((.....))))).....)))))))..)))). ( -38.60) >DroMoj_CAF1 23830 120 + 1 ACUCCGGCAAGUCCACCACCACCGGUCACUUGAUCUACAAGUGCGGUGGUAUCGACAAGCGUACCAUUGAGAAGUUCGAGAAGGAAGCCCAGGAGAUGGGCAAGGGCUCGUUCAAGUACG (((..(((.(((((.(((((((((..((((((.....))))))))))))).((((....((......))......))))...))..(((((.....)))))..))))).)))..)))... ( -42.40) >DroPer_CAF1 25282 120 + 1 AUUCUGGCAAGUCCACGACCACCGGUCACUUGAUCUACAAGUGCGGUGGUAUCGACAAGCGUACCAUUGAGAAGUUCGAGAAGGAGGCUCAGGAGAUGGGCAAGGGCUCCUUCAAGUACG ......((..(((....(((((((..((((((.....)))))))))))))...)))..))((((..(((((...)))))((((((((((((.....))))).....)))))))..)))). ( -43.60) >consensus ACUCUGGCAAGUCCACCACCACCGGUCACUUGAUCUACAAGUGCGGUGGUAUCGACAAGCGUACCAUUGAGAAGUUCGAGAAGGAAGCUCAGGAAAUGGGCAAGGGCUCCUUCAAGUACG ......((..(((....(((((((..((((((.....)))))))))))))...)))..))((((..(((((...)))))(((.((.(((((.....)))))......)).)))..)))). (-34.62 = -34.23 + -0.39)
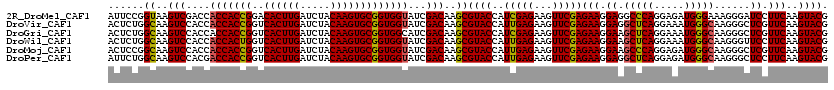
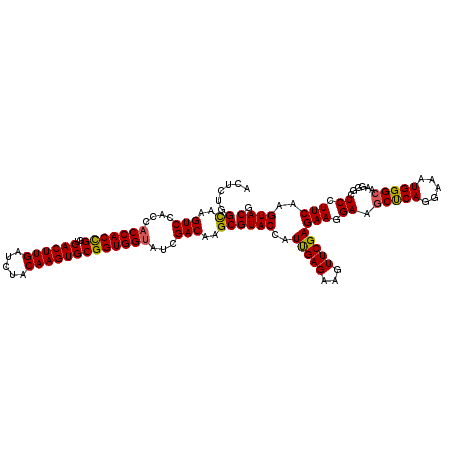
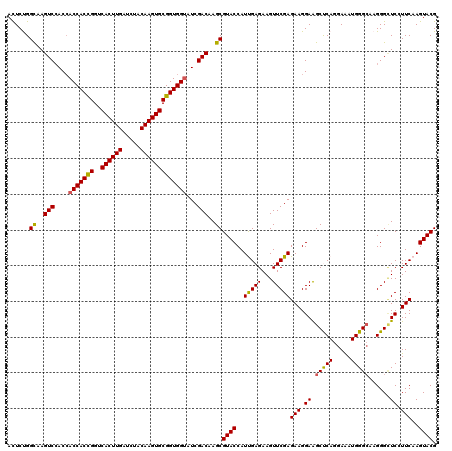
| Location | 7,409,570 – 7,409,690 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -39.57 |
| Consensus MFE | -34.06 |
| Energy contribution | -33.45 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7409570 120 + 20766785 GUGCGGUGGUAUCGACAAGCGUACCAUCGAGAAGUUCGAGAAGGAGGCCCAGGAGAUGGGAAAGGGAUCCUUCAAGUACGCCUGGGUUUUGGAUAAGUUGAAGGCUGAGCGCGAGCGUGG ...((((((((.((.....))))))))))....((((..((((((..((((.....)))).......))))))......((((.((((((.((....)).)))))).)).)))))).... ( -41.30) >DroVir_CAF1 23911 120 + 1 GUGCGGUGGUAUCGACAAGCGUACCAUUGAGAAGUUCGAGAAGGAGGCUCAGGAAAUGGGCAAGGGCUCGUUCAAGUACGCAUGGGUUUUGGAUAAAUUGAAGGCCGAGCGUGAGCGUGG ...((((((((.((.....))))))))))....((((..(((.((((((((.....))))).....))).)))....((((.(.((((((.((....)).)))))).))))))))).... ( -37.70) >DroGri_CAF1 24039 120 + 1 GUGCGGUGGCAUCGACAAGCGUACCAUCGAGAAGUUCGAGAAGGAAGCUCAGGAAAUGGGCAAGGGCUCGUUCAAGUACGCAUGGGUUUUGGAUAAAUUGAAGGCCGAGCGUGAGCGUGG ..(((.(.((.((..((.((((((....(((...(((.....)))..)))..(((.(((((....))))))))..)))))).))((((((.((....)).))))))))..)).).))).. ( -36.10) >DroWil_CAF1 27549 120 + 1 GUGCGGUGGUAUCGACAAGCGUACCAUUGAGAAGUUCGAGAAGGAAGCUCAGGAAAUGGGCAAGGGUUCCUUCAAGUACGCAUGGGUUUUGGAUAAAUUGAAGGCCGAGCGUGAGCGUGG ...((((((((.((.....))))))))))....((((..((((((((((((.....))))).....)))))))....((((.(.((((((.((....)).)))))).))))))))).... ( -40.40) >DroYak_CAF1 24122 120 + 1 GUGCGGUGGUAUCGACAAGCGUACCAUCGAGAAGUUCGAGAAGGAAGCCCAGGAGAUGGGCAAGGGAUCCUUCAAGUACGCCUGGGUUUUGGAUAAGUUGAAGGCUGAGCGCGAGCGUGG ...((((((((.((.....))))))))))....((((..((((((.(((((.....)))))......))))))......((((.((((((.((....)).)))))).)).)))))).... ( -42.70) >DroMoj_CAF1 23870 120 + 1 GUGCGGUGGUAUCGACAAGCGUACCAUUGAGAAGUUCGAGAAGGAAGCCCAGGAGAUGGGCAAGGGCUCGUUCAAGUACGCAUGGGUUUUGGAUAAAUUGAAGGCCGAGCGUGAGCGUGG (((((((((((.((.....))))))))......(..((((......(((((.....))))).....))))..).....))))).((((((.((....)).))))))..((....)).... ( -39.20) >consensus GUGCGGUGGUAUCGACAAGCGUACCAUCGAGAAGUUCGAGAAGGAAGCCCAGGAAAUGGGCAAGGGCUCCUUCAAGUACGCAUGGGUUUUGGAUAAAUUGAAGGCCGAGCGUGAGCGUGG ...((((((((.((.....))))))))))....(((((.(((.((.(((((.....)))))......)).))).....(((.(.((((((.((....)).)))))).))))))))).... (-34.06 = -33.45 + -0.61)

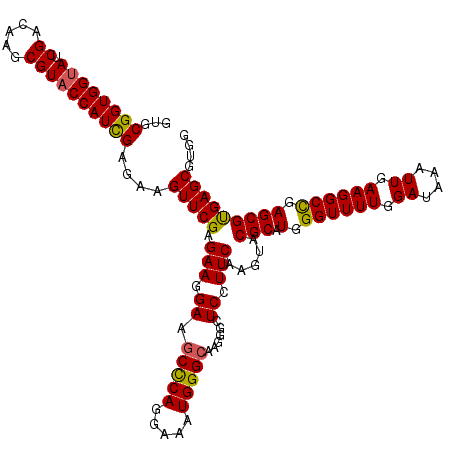
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:29 2006