
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,401,277 – 7,401,386 |
| Length | 109 |
| Max. P | 0.690467 |

| Location | 7,401,277 – 7,401,386 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -20.98 |
| Energy contribution | -20.73 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7401277 109 + 20766785 UGUCUUCAGAUUCGUUCGCAGUAAGCUCCAGCAACCGCUGGGCGCCAAGCCGCAGCUUGCUGACGUUCACCUCCAUCUCAACAGGAUCCACUCAACUGAUUUUUA--AGUG .....((((....(..((((((((((((((((....)))))(((......)))))))))))).))..)...(((.........))).........))))......--.... ( -31.90) >DroSec_CAF1 17461 109 + 1 UGUCUUCAGAUUCGUUCGCAGUAAGUUCCAGCAACCGCUGGGCGCCUAGCCGCAGCUUGCUGACGUUCACUUCCAUUUCAACAGGAUCCACUCAACUGAUUUUUA--AGUG .....((((....(..((((((((((((((((....)))))(((......)))))))))))).))..)...(((.........))).........))))......--.... ( -29.60) >DroSim_CAF1 8889 109 + 1 UGUCUUCAGAUUCGUUCGCAGUAAGUUCCAGCAACCGCUGGGCGCCUAGCCGCAGCUUGCUGACGUUCACCUCCAUUUCAACAGGAUCCACUCAACUGAUUUUUA--AGGG .....((((....(..((((((((((((((((....)))))(((......)))))))))))).))..)...(((.........))).........))))......--.... ( -29.50) >DroEre_CAF1 17241 97 + 1 UGUCUUCAGACUCGUUUGCAGUAAGUUCCAGCAACCGCUGAGCGCCAAGCCGCAGCUUACUGGCAUUUACUUCCAUCUCAACAGAAUCCACUCAAUU-------------- .(((....)))..((.(((((((((((.((((....)))).(((......))))))))))).)))...))...........................-------------- ( -21.60) >DroYak_CAF1 15825 97 + 1 UGUCUUCGGACUCGUUUGCAGUAAGUUCCAGCAACCGCUGGGCGCCAAGCCGCAGCUUACUGGCAUUCACUUCCAUCUCAACAGGAUCCACUCAAUA-------------- .(((....)))..(..((((((((((((((((....)))))(((......))))))))))).)))..)...(((.........)))...........-------------- ( -29.90) >DroPer_CAF1 18137 111 + 1 UGUCUUGGGGCUCGUUGGCAGUCAACUGCAGAAGUCGCUGUGCACAGAAUCGUAGCUUGCUGGCAUUUACCUCCAUUGCCUUGGUAGCCGGUCAACUGUAUUCGGAUACUG .((..(((((.....((.(((.(((((((.((.....(((....)))..)))))).)))))).)).....)))))..))...((((.((((..........)))).)))). ( -28.30) >consensus UGUCUUCAGACUCGUUCGCAGUAAGUUCCAGCAACCGCUGGGCGCCAAGCCGCAGCUUGCUGACAUUCACCUCCAUCUCAACAGGAUCCACUCAACUGAUUUUUA__AGUG .(((....)))..(..((((((((((((((((....)))))(((......)))))))))))).))..)........................................... (-20.98 = -20.73 + -0.25)

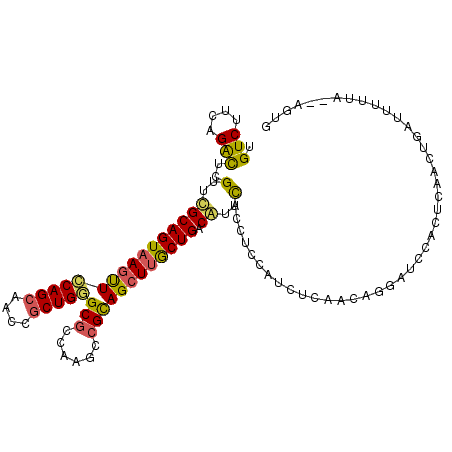
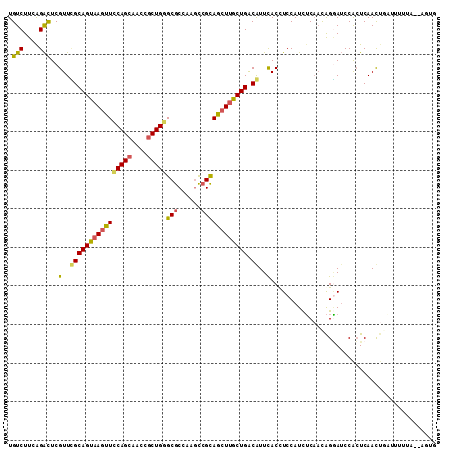
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:23 2006