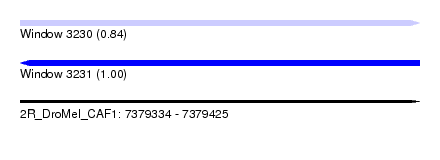
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,379,334 – 7,379,425 |
| Length | 91 |
| Max. P | 0.999508 |

| Location | 7,379,334 – 7,379,425 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -24.96 |
| Consensus MFE | -20.84 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7379334 91 + 20766785 GUUUUCGAUGCCCAGUGUGGACAUAGAUUCCGUUUCCGAUUCGGGUUCGUAUCCGAAAUUCGGCUGAUGAAAACGAAACUGAAAACUGC--AC ((((((((((.((.....)).)))......((((.(((((((((((....)))))))..))))..))))..........)))))))...--.. ( -23.40) >DroSec_CAF1 2107 91 + 1 GUUUUCGAUGCCCAGUGUGGACAUGGAUUCCGUAUCCGAUUCGGAUUCGUAUCCGAAAUUCGGCUGAUGAAAACGAAACUGAAAACUGC--AC (((((((..(((.(((.((((.((((...)))).)))).(((((((....)))))))))).)))...)))))))...............--.. ( -26.20) >DroSim_CAF1 2431 91 + 1 GUUUUCGAUGCCCAGUGUGGACAUGGAUUCCGUAUCCGAUUCAGAUUCGUAUCCGAAAUUCGGCUGAAGAAAACGAAACUAAAAACUGC--AC ((((((...(((.(((.((((.((((...)))).)))).(((.(((....))).)))))).)))....))))))...............--.. ( -20.10) >DroEre_CAF1 2009 88 + 1 GUUUUCGAUGCCA---GUGGACAUGGAUUCCGUUUCCGAUUCGGAUUCGAAUCCAAAAUUCGGCUGAUGGAAACGAAACCGAAAACUGC--AC (((((((....((---((.((..(((((((.(..((((...))))..))))))))....)).)))).((....))....)))))))...--.. ( -29.80) >DroYak_CAF1 2134 90 + 1 GUUUUCGAUGCCA---GUGGACAUGGAUUCCGUUUCCGAUUCGGAUUCGAAUCCAAAAUUCGGCUGAUGAAAACGAAACUGAAAACUGAAAAC (((((((....((---((.((..(((((((.(..((((...))))..))))))))....)).)))).)))))))................... ( -25.30) >consensus GUUUUCGAUGCCCAGUGUGGACAUGGAUUCCGUUUCCGAUUCGGAUUCGUAUCCGAAAUUCGGCUGAUGAAAACGAAACUGAAAACUGC__AC (((((((..(((.....((((.((((...)))).)))).(((((((....)))))))....)))...)))))))................... (-20.84 = -21.52 + 0.68)

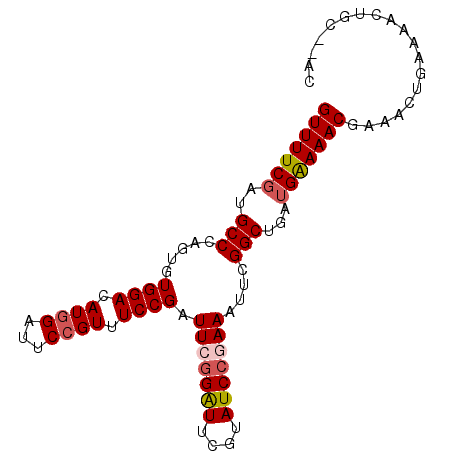
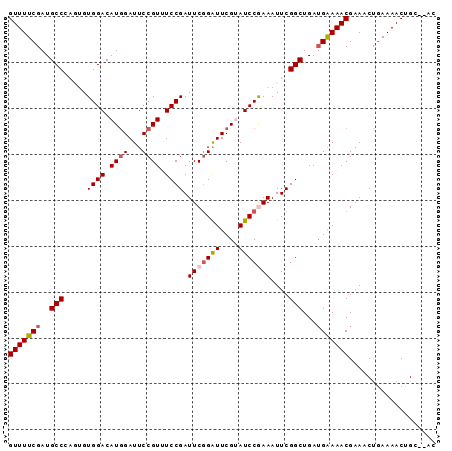
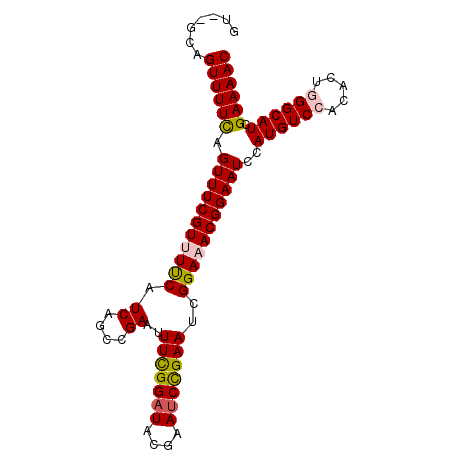
| Location | 7,379,334 – 7,379,425 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -24.54 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7379334 91 - 20766785 GU--GCAGUUUUCAGUUUCGUUUUCAUCAGCCGAAUUUCGGAUACGAACCCGAAUCGGAAACGGAAUCUAUGUCCACACUGGGCAUCGAAAAC ..--...((((((.(((((((((((.((....))..(((((........)))))..)))))))))))..((((((.....)))))).)))))) ( -27.10) >DroSec_CAF1 2107 91 - 1 GU--GCAGUUUUCAGUUUCGUUUUCAUCAGCCGAAUUUCGGAUACGAAUCCGAAUCGGAUACGGAAUCCAUGUCCACACUGGGCAUCGAAAAC ..--...((((((......(..(((.....((((..(((((((....))))))))))).....)))..)((((((.....)))))).)))))) ( -28.40) >DroSim_CAF1 2431 91 - 1 GU--GCAGUUUUUAGUUUCGUUUUCUUCAGCCGAAUUUCGGAUACGAAUCUGAAUCGGAUACGGAAUCCAUGUCCACACUGGGCAUCGAAAAC ..--...((((((......(..((((....((((..(((((((....)))))))))))....))))..)((((((.....)))))).)))))) ( -24.10) >DroEre_CAF1 2009 88 - 1 GU--GCAGUUUUCGGUUUCGUUUCCAUCAGCCGAAUUUUGGAUUCGAAUCCGAAUCGGAAACGGAAUCCAUGUCCAC---UGGCAUCGAAAAC ..--...((((((((((((((((((.((....))......((((((....)))))))))))))))))).(((((...---.))))).)))))) ( -30.10) >DroYak_CAF1 2134 90 - 1 GUUUUCAGUUUUCAGUUUCGUUUUCAUCAGCCGAAUUUUGGAUUCGAAUCCGAAUCGGAAACGGAAUCCAUGUCCAC---UGGCAUCGAAAAC .......((((((.(((((((((((.((....))......((((((....)))))))))))))))))..(((((...---.))))).)))))) ( -24.50) >consensus GU__GCAGUUUUCAGUUUCGUUUUCAUCAGCCGAAUUUCGGAUACGAAUCCGAAUCGGAAACGGAAUCCAUGUCCACACUGGGCAUCGAAAAC .......((((((.(((((((((((.((....))..(((((((....)))))))..)))))))))))..((((((.....)))))).)))))) (-24.54 = -24.82 + 0.28)

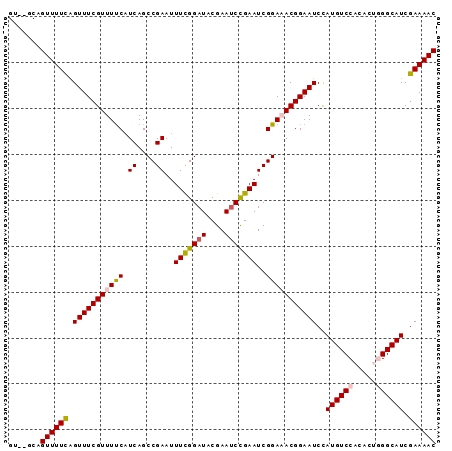
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:14 2006