
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,375,893 – 7,375,997 |
| Length | 104 |
| Max. P | 0.851211 |

| Location | 7,375,893 – 7,375,997 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.32 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -18.06 |
| Energy contribution | -17.95 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7375893 104 - 20766785 CUUGCCACCAAUGCAAGUACUA----------------CUUACCUCAACACCCACCGCCUGCUUUUCACCCUCGGAGGAGACGCCGUCGGCUCCCUCGCUGUCCUCGGUGGGGUUUCCCU (((((.......))))).....----------------........(((.(((((((...((.((((..(....)..)))).)).(.((((......)))).)..))))))))))..... ( -29.40) >DroVir_CAF1 12244 81 - 1 --------------------AG----------------CUUACCUCAACUUCGACGGCCUGUUUUUCGCCCUCUGACGAGACGCCAUCGGCGCCCUCGUUAUCCUCGGAGGG---CCCUU --------------------((----------------(...((((......)).))...)))....(((((((((((((.((((...))))..))))......))))))))---).... ( -26.70) >DroSec_CAF1 7235 104 - 1 CUUGCCACCAAGGCAAGUACUA----------------CUUACCUCAACACCCACCGCCUGCUUCUCACCCUCGGAGGAGACGCCGUCGGCUCCCUCGCUGUCCUCGGUGGGGUUUCCCU ((((((.....)))))).....----------------........(((.(((((((...((.((((..(....)..)))).)).(.((((......)))).)..))))))))))..... ( -36.30) >DroSim_CAF1 7531 104 - 1 CUUGCCACCAAGGCAAGUACUA----------------CUUACCUCAACACCCACCGCCUGCUUCUCACCCUCGGAGGAGACGCCGUCGGCUCCCUCGCUGUCCUCGGUGGGGUUUCCCU ((((((.....)))))).....----------------........(((.(((((((...((.((((..(....)..)))).)).(.((((......)))).)..))))))))))..... ( -36.30) >DroYak_CAF1 10395 108 - 1 ------------GAAAGUGUUACUUUGCUCAAGGAGUCCUUACCUCAACACCCACCGCCUGCUUCUCACCCUCGGAGGAAACGCCGUCGGCUCCUUCGUUGUCCUCGGUGGGGUUACCCU ------------....(((..(((((......)))))...)))...(((.(((((((...((..........(((((((..((....))..)))))))..))...))))))))))..... ( -28.20) >DroAna_CAF1 7410 101 - 1 CUUAUCCCUG---A----------------AAUGUGUCCUUACCUCCACACCCACGGCCUGCUUCUCACCCUCCGACGAUACGCCAUCAGUGCCUUCGCUGUCCUCCGUGGGGUUGCCCU ..........---.----------------..((((..........))))(((((((...((.((((.......)).))...))...(((((....)))))....)))))))........ ( -19.40) >consensus CUUGCCACCA__GCAAGUACUA________________CUUACCUCAACACCCACCGCCUGCUUCUCACCCUCGGAGGAGACGCCGUCGGCUCCCUCGCUGUCCUCGGUGGGGUUUCCCU ..................................................(((((((...((.((((..(....)..)))).))...((((......))))....)))))))........ (-18.06 = -17.95 + -0.11)
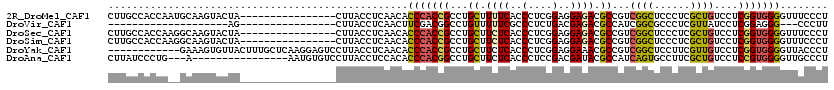
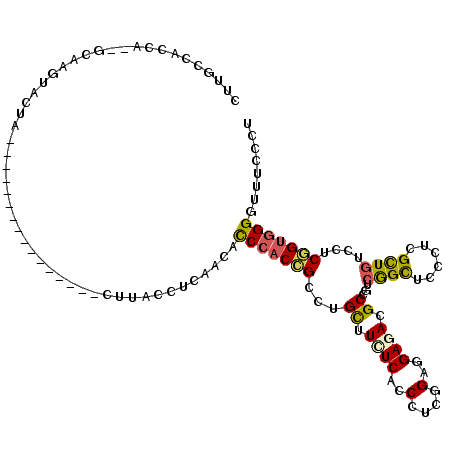
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:12 2006