
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,368,844 – 7,369,000 |
| Length | 156 |
| Max. P | 0.910211 |

| Location | 7,368,844 – 7,368,934 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 65.33 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -11.99 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7368844 90 - 20766785 AAGAAGCGGUAUCUGAGCCGGUAAACGCCGCAAAGUUGGA------------GGUAAAAGCAUAC-GCAUGCAA--AUAAAGGGUUAUUUGCAAAAUAUGGCGCA .....(((((((((..(((....(((........)))...------------)))...)).))))-...(((((--((((....)))))))))........))). ( -21.30) >DroSim_CAF1 3093 102 - 1 AAGAAGCGGAAUCUGAGCCGGCAAACGCCGAACAGUUGGAGCCCCAUAUGUAGGUACAAGCAUACCGCAUGCAA--AUAAAA-GUUAUAUGUGAAAUAUGGCGCA .....((((.((((..((((((....)))..(((..(((....)))..))).)))...)).)).))))......--......-(((((((.....)))))))... ( -25.90) >DroEre_CAF1 195 79 - 1 AAUUAGUGCGAUCUGAGCCGGCAAAUGCCGAAGAU---------------UGG---AAAACGU-----AUGCAU--AUUGAA-GUUAUAUGCGAACUGUAGAGCA .....((.((((((....((((....)))).))))---------------)).---.....((-----.(((((--((....-...))))))).))......)). ( -19.80) >DroYak_CAF1 3162 97 - 1 AAGUAGUACAAUCUGAGCCGGCAAACGCCGAAAACAUAACGCCC--UCAGUGGGUACAAGCAU-----AUGCAUAUAUUUAA-GUUAUACGCGAAAUGCUGUGCA .....((((.........((((....))))..........((((--.....))))...(((((-----.(((.((((.....-..)))).)))..))))))))). ( -24.60) >consensus AAGAAGCGCAAUCUGAGCCGGCAAACGCCGAAAAGUUGGA__________UGGGUAAAAGCAU_____AUGCAA__AUAAAA_GUUAUAUGCGAAAUAUGGCGCA .....(((((((....((((((....)))).............................((((.....))))..................)).....))))))). (-11.99 = -12.30 + 0.31)

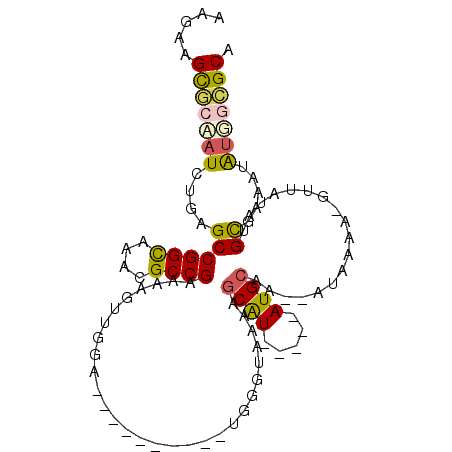
| Location | 7,368,894 – 7,369,000 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.96 |
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -8.78 |
| Energy contribution | -8.90 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7368894 106 + 20766785 UCCAACUUUGCGGCGUUUACCGGCUCAGAUACCGCUUCUUGUAA--------------UAAGCAAUCCGCGACUGUAUGGUUUCUAGCGUGGUAAACACUCUUGCGCCAUUUACAUAUUU ...........((((((((((((((.(((.(((((((.......--------------.)))).....((....))..))).)))))).)))))))).(....).)))............ ( -25.80) >DroSec_CAF1 226 105 + 1 UUCAACUGUUCGGCGUUUGCCGGCUCAGAUUCCGCUUCUUGUAA--------------UAAGCAAG-CGCUAUUGUAUAGUUUCUAGCGUGCUAAAAACUCUUGCACCAUUUAUAUAUUU .....(((.(((((....)))))..)))....(((((((((...--------------)))).)))-))....((((.(((((.(((....))).)))))..)))).............. ( -23.30) >DroSim_CAF1 3155 104 + 1 UCCAACUGUUCGGCGUUUGCCGGCUCAGAUUCCGCUUCUUGUAA--------------UAAGCAAG-CGCUAUUGUAUAGUUUCUAGCGUGCUAAACACUCUUGCGCCAUUUAUAUAU-U .....(((.(((((....)))))..))).....((((.......--------------.))))..(-(((...(((.((((.........)))).))).....))))...........-. ( -22.40) >DroEre_CAF1 239 114 + 1 -----AUCUUCGGCAUUUGCCGGCUCAGAUCGCACUAAUUGUACAUCUGUACUUUUAGUUGGCAAU-CGCAAAUGUCGAUUCUCAUGCUUGAUAAACAGUCUUGAGGCAUUUAUAUAGUU -----....(((((((((((.......(((.(((((((..((((....))))..)))))..)).))-)))))))))))).(((((.(((........)))..)))))............. ( -30.41) >DroYak_CAF1 3219 109 + 1 GUUAUGUUUUCGGCGUUUGCCGGCUCAGAUUGUACUACUUGUAC-----UACUUGUA-----CAAU-CGCAAUUAUAAAUUUUCUUGCUGUCUAAAUACUCUUGAGCCAUUUAUAUAGUU .((((((....(((....)))((((((((((((((.........-----.....)))-----))))-)((((..(......)..))))..............))))))....)))))).. ( -27.54) >consensus UCCAACUGUUCGGCGUUUGCCGGCUCAGAUUCCGCUUCUUGUAA______________UAAGCAAU_CGCAAUUGUAUAGUUUCUAGCGUGCUAAACACUCUUGCGCCAUUUAUAUAUUU ...........(((....)))(((.(((.....((.....))..........................((((..(......)..)))).............))).)))............ ( -8.78 = -8.90 + 0.12)

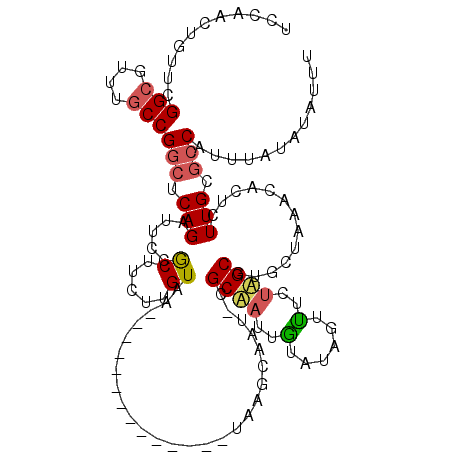

| Location | 7,368,894 – 7,369,000 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.96 |
| Mean single sequence MFE | -25.99 |
| Consensus MFE | -10.57 |
| Energy contribution | -10.69 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7368894 106 - 20766785 AAAUAUGUAAAUGGCGCAAGAGUGUUUACCACGCUAGAAACCAUACAGUCGCGGAUUGCUUA--------------UUACAAGAAGCGGUAUCUGAGCCGGUAAACGCCGCAAAGUUGGA .............(((.....(((((((((..((((((.(((....((((...))))((((.--------------.......))))))).))).))).))))))))))))......... ( -27.00) >DroSec_CAF1 226 105 - 1 AAAUAUAUAAAUGGUGCAAGAGUUUUUAGCACGCUAGAAACUAUACAAUAGCG-CUUGCUUA--------------UUACAAGAAGCGGAAUCUGAGCCGGCAAACGCCGAACAGUUGAA ............(((...(((((((((((....))))))))..........((-(((.(((.--------------....))))))))...)))..)))(((....)))........... ( -25.20) >DroSim_CAF1 3155 104 - 1 A-AUAUAUAAAUGGCGCAAGAGUGUUUAGCACGCUAGAAACUAUACAAUAGCG-CUUGCUUA--------------UUACAAGAAGCGGAAUCUGAGCCGGCAAACGCCGAACAGUUGGA .-..........(((...(((((.(((((....))))).))..........((-(((.(((.--------------....))))))))...)))..)))(((....)))........... ( -23.30) >DroEre_CAF1 239 114 - 1 AACUAUAUAAAUGCCUCAAGACUGUUUAUCAAGCAUGAGAAUCGACAUUUGCG-AUUGCCAACUAAAAGUACAGAUGUACAAUUAGUGCGAUCUGAGCCGGCAAAUGCCGAAGAU----- ..............((((.(.((........))).))))..(((.((((((((-(((((..(((((..((((....))))..))))))))))).......))))))).)))....----- ( -28.01) >DroYak_CAF1 3219 109 - 1 AACUAUAUAAAUGGCUCAAGAGUAUUUAGACAGCAAGAAAAUUUAUAAUUGCG-AUUG-----UACAAGUA-----GUACAAGUAGUACAAUCUGAGCCGGCAAACGCCGAAAACAUAAC ............((((((..............((((............))))(-((((-----(((.....-----.........))))))))))))))(((....)))........... ( -26.44) >consensus AAAUAUAUAAAUGGCGCAAGAGUGUUUAGCACGCUAGAAACUAUACAAUAGCG_AUUGCUUA______________UUACAAGAAGCGGAAUCUGAGCCGGCAAACGCCGAAAAGUUGAA ............(((...(((...........((((............))))....(((((......................)))))...)))..)))(((....)))........... (-10.57 = -10.69 + 0.12)

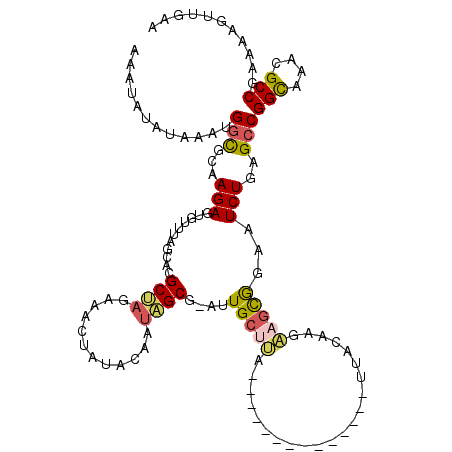
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:10 2006