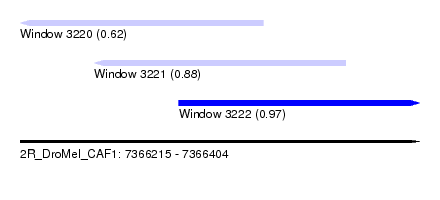
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,366,215 – 7,366,404 |
| Length | 189 |
| Max. P | 0.970290 |

| Location | 7,366,215 – 7,366,330 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 97.68 |
| Mean single sequence MFE | -40.87 |
| Consensus MFE | -38.07 |
| Energy contribution | -37.63 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7366215 115 - 20766785 UCCCACCUGGCUGCUUGCUGGCAUAGUAGUUCAGGUGGUUCUUGGACUCCUCCAGGAACAGAUGCUCGAUAAACUUGUCGCUCAGCUCGUGGCCUGCACCCACAUCGCAGGUGUC ...((((((((((((((....)).))))).)))))))(((((((((....))))))))).((.((((((((....)))))...)))))...((((((.........))))))... ( -45.00) >DroSim_CAF1 373 115 - 1 UCCCACCUGGCUGCUUGCUGGCAUAGUAGUUCAGGUGGUUCUUGGACUCCUCCAGGAACAGAUGCUCGAUAAACUUGUCGCUCAGCUCGUGGCCCGCACCCACAUCGCAGGUGUC ...((((((((((...((.((((.(((...((..((.(((((((((....)))))))))....))..))...))))))))).))))..((((.......))))....)))))).. ( -41.30) >DroYak_CAF1 393 115 - 1 UCCCACCUGGCUGCUUACUGGCAUAGUAGUUCAGGUGGUUUUUGGACUCCUCCAGGAACAGAUGCUCGAUAAACUUGUCACUCAGCUCGUGGCCCGCACCCACAUCGCAGGUGUC ...((((((((((.....(((((.(((...((..((.(((((((((....)))))))))....))..))...))))))))..))))..((((.......))))....)))))).. ( -36.30) >consensus UCCCACCUGGCUGCUUGCUGGCAUAGUAGUUCAGGUGGUUCUUGGACUCCUCCAGGAACAGAUGCUCGAUAAACUUGUCGCUCAGCUCGUGGCCCGCACCCACAUCGCAGGUGUC ...((((((((((.....(((((.(((...((..((.(((((((((....)))))))))....))..))...))))))))..))))..((((.......))))....)))))).. (-38.07 = -37.63 + -0.44)

| Location | 7,366,250 – 7,366,369 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.81 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -30.92 |
| Energy contribution | -30.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7366250 119 - 20766785 GGGAUUUAUUAUUUGGAGUUUUC-CUCGAUGAAAUAAUACUCCCACCUGGCUGCUUGCUGGCAUAGUAGUUCAGGUGGUUCUUGGACUCCUCCAGGAACAGAUGCUCGAUAAACUUGUCG (((((((.......(((((((((-......))))....)))))((((((((((((((....)).))))).)))))))(((((((((....))))))))))))).)))((((....)))). ( -39.30) >DroSim_CAF1 408 119 - 1 GGGCCUUAUUAUUUGAAAUUUUC-CUCGAUGAAAUAAUACUCCCACCUGGCUGCUUGCUGGCAUAGUAGUUCAGGUGGUUCUUGGACUCCUCCAGGAACAGAUGCUCGAUAAACUUGUCG ((((..((((((((...(((...-...))).))))))))....((((((((((((((....)).))))).)))))))(((((((((....)))))))))....))))((((....)))). ( -37.40) >DroYak_CAF1 428 119 - 1 GGCAUUUAUUAUUUGUAAUUUUUUUUUAACGAAAU-AUAUUCCCACCUGGCUGCUUACUGGCAUAGUAGUUCAGGUGGUUUUUGGACUCCUCCAGGAACAGAUGCUCGAUAAACUUGUCA (((((((............................-.......((((((((((((.........))))).)))))))(((((((((....)))))))))))))))).((((....)))). ( -32.90) >consensus GGGAUUUAUUAUUUGAAAUUUUC_CUCGAUGAAAUAAUACUCCCACCUGGCUGCUUGCUGGCAUAGUAGUUCAGGUGGUUCUUGGACUCCUCCAGGAACAGAUGCUCGAUAAACUUGUCG .........((((((...........................(((((((((((((.........))))).))))))))((((((((....))))))))))))))...((((....)))). (-30.92 = -30.70 + -0.22)

| Location | 7,366,290 – 7,366,404 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.24 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -30.09 |
| Energy contribution | -30.43 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7366290 114 + 20766785 AACCACCUGAACUACUAUGCCAGCAAGCAGCCAGGUGGGAGUAUUAUUUCAUCGAG-GAAAACUCCAAAUAAUAAAUCCCAUUUGGUUACUUAGCCCAGGACAAGUCAGCGCAGG .....((((..(((((.((((.(((((.((((((((((((.((((((((....(((-.....))).))))))))..)))))))))))).))).))...)).))))).))..)))) ( -38.70) >DroSim_CAF1 448 114 + 1 AACCACCUGAACUACUAUGCCAGCAAGCAGCCAGGUGGGAGUAUUAUUUCAUCGAG-GAAAAUUUCAAAUAAUAAGGCCCAUUUGGUUACUUAGCCCAGGACAAGUCAGCGCAGG .....((((..(((((.((((.(((((.(((((((((((..((((((((....(((-(....))))))))))))...))))))))))).))).))...)).))))).))..)))) ( -35.00) >DroYak_CAF1 468 114 + 1 AACCACCUGAACUACUAUGCCAGUAAGCAGCCAGGUGGGAAUAU-AUUUCGUUAAAAAAAAAUUACAAAUAAUAAAUGCCUUUUGGCUCCUUAGCCCAGGACAAGUCGGCGCAGG ..(((((((........(((......)))..)))))))(((...-..)))...........................(((....))).(((..(((...((....)))))..))) ( -23.00) >consensus AACCACCUGAACUACUAUGCCAGCAAGCAGCCAGGUGGGAGUAUUAUUUCAUCGAG_GAAAAUUACAAAUAAUAAAUCCCAUUUGGUUACUUAGCCCAGGACAAGUCAGCGCAGG .....((((..(((((.((((.(((((.(((((((((((..((((((((.................))))))))...))))))))))).))).))...)).))))).))..)))) (-30.09 = -30.43 + 0.34)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:06 2006