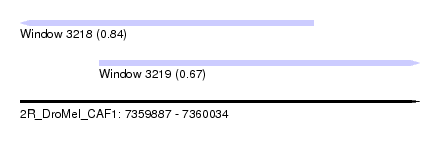
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,359,887 – 7,360,034 |
| Length | 147 |
| Max. P | 0.841006 |

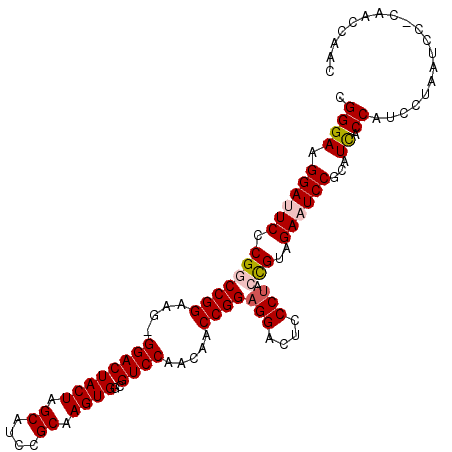
| Location | 7,359,887 – 7,359,995 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 88.27 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -26.86 |
| Energy contribution | -27.54 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7359887 108 - 20766785 CGGGAAGGAAUCCCGUCCGGUAG-GGACUACUAGCAUUCGCAAGUGACGUCCAGCAACCGGAGGACCCCCAACUGUAGAAUCCGCAUCACCAUCCUAAUCC-CAACAAAC .((((((((...((.((((((..-((((((((.((....)).))))..))))....)))))))).........(((.......)))......))))..)))-)....... ( -32.30) >DroSec_CAF1 6845 108 - 1 CGGGAAGGAUUCCCGGCCGGAAG-GGACUACUAGCAUCCGCAAGUGGCGUCCAACAACCGGAGGACUCCCUACCGUAGAAUCCGCAUCACCAUCCUAAUCC-AAAGCAAC .((((.((((((...((.((.((-((((((((.((....)).))))).((((..(....)..))))))))).)))).))))))...)).))..........-........ ( -34.70) >DroSim_CAF1 13292 108 - 1 CGGGAAGGAUUCCCGGCCGGAAG-GGACUACUAGCAUCCGCAAGUGGCGUCCAACAACCGGAGGACUCCCUACCGUAGAAUCCGCAUCACCAUCCUAAUCC-CAACCAAC .((((.((((((...((.((.((-((((((((.((....)).))))).((((..(....)..))))))))).)))).))))))(......).......)))-)....... ( -36.00) >DroEre_CAF1 6862 109 - 1 CGGGAAGGAUUCCCGUCCGGAAGGGGACUACUAGCAUCCGCAAGUGGCGUCCAAUAACCGGAGGUUUCCCUACCGAGGAAUCCGCAUCACCAUUGUAGAAC-CAAUCAAC .((((.(((((((....(((.(((((((((((.((....)).)))))(.(((.......))).).)))))).))).)))))))...)).))..........-........ ( -36.30) >DroYak_CAF1 6231 110 - 1 CGGGAAGGAUUCCCGGCCGGAAGGGGACUACUAGCAUCCGCAAGUGGCGUCCAGCAACCGGAGGUCUCCCUUCCGAAGAUUCCGCAUUACCAUCCUAUAACCCAACCAAC .(((.(((((.....(((((((((((...(((.((....)).)))(((.(((.......))).))))))))))))..(....)))......)))))....)))....... ( -35.30) >consensus CGGGAAGGAUUCCCGGCCGGAAG_GGACUACUAGCAUCCGCAAGUGGCGUCCAACAACCGGAGGACUCCCUACCGUAGAAUCCGCAUCACCAUCCUAAUCC_CAACCAAC .((((.((((((.(((((((....((((((((.((....)).))))..)))).....))))(((....))).)))..))))))...)).))................... (-26.86 = -27.54 + 0.68)

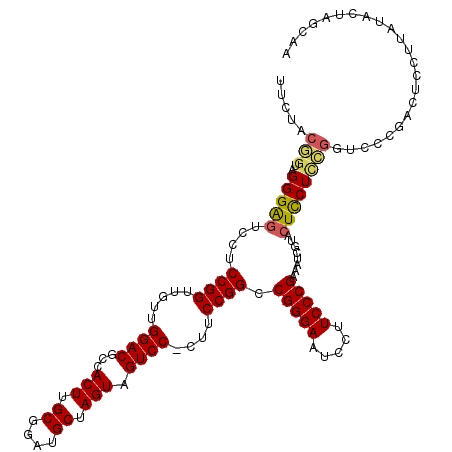
| Location | 7,359,916 – 7,360,034 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.92 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -29.42 |
| Energy contribution | -29.54 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7359916 118 + 20766785 UUCUACAGUUGGGGGUCCUCCGGUUGCUGGACGUCACUUGCGAAUGCUAGUAGUCC-CUACCGGACGGGAUUCCUUCCCGAAAUCUUACUCCUCUGGUCCCGACUCCUUAUACCAGCAA .......((((((((.((((((((....((((...(((.((....)).))).))))-..))))))(((((.....)))))...............)).)))(....).....))))).. ( -40.80) >DroSec_CAF1 6874 118 + 1 UUCUACGGUAGGGAGUCCUCCGGUUGUUGGACGCCACUUGCGGAUGCUAGUAGUCC-CUUCCGGCCGGGAAUCCUUCCCGAAAUCGUACUCCUCCGGUCCCGACUACUUAUACUAGCAA .....(((.(((....)))))).(((((((.(((.....)))......(((((((.-...((((.(((((.....))))).....(....)..))))....)))))))....))))))) ( -35.70) >DroSim_CAF1 13321 118 + 1 UUCUACGGUAGGGAGUCCUCCGGUUGUUGGACGCCACUUGCGGAUGCUAGUAGUCC-CUUCCGGCCGGGAAUCCUUCCCGAAAUCGUACUCCUUCGGUCCCGACUCCUUAUACUAGCAA ..(((..((((((((((.((((((((..((((...(((.((....)).))).))))-....))))))))........(((((..........)))))....))))))))))..)))... ( -39.70) >DroEre_CAF1 6891 107 + 1 UUCCUCGGUAGGGAAACCUCCGGUUAUUGGACGCCACUUGCGGAUGCUAGUAGUCCCCUUCCGGACGGGAAUCCUUCCCGAAAUCGUACUCCU------------CCUAGUACUAGCAA .(((((((.(((....))))))).....)))(((.....)))..((((((((((((......))))((((.....))))..............------------.....)))))))). ( -37.20) >DroYak_CAF1 6261 119 + 1 AUCUUCGGAAGGGAGACCUCCGGUUGCUGGACGCCACUUGCGGAUGCUAGUAGUCCCCUUCCGGCCGGGAAUCCUUCCCGAUCUCGUACUUCUCCGGUUCCGACUCCAAGUACUAGCUA .....((((((((.(((.......((((((.(((.....)))....)))))))))))))))))(((((((.....))))).....((((((...((....)).....))))))..)).. ( -41.11) >consensus UUCUACGGUAGGGAGUCCUCCGGUUGUUGGACGCCACUUGCGGAUGCUAGUAGUCC_CUUCCGGCCGGGAAUCCUUCCCGAAAUCGUACUCCUCCGGUCCCGACUCCUUAUACUAGCAA .....(((..(((((....((((.....((((...(((.((....)).))).))))....)))).(((((.....)))))........))))))))....................... (-29.42 = -29.54 + 0.12)

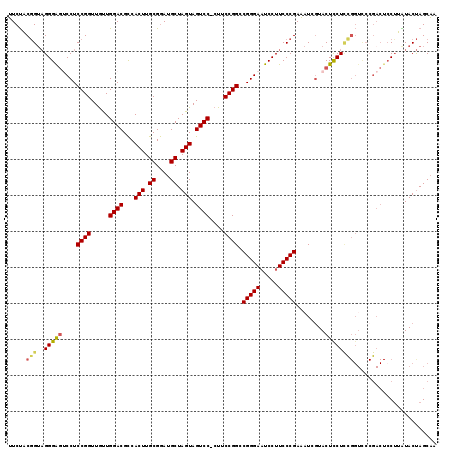
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:03 2006