
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
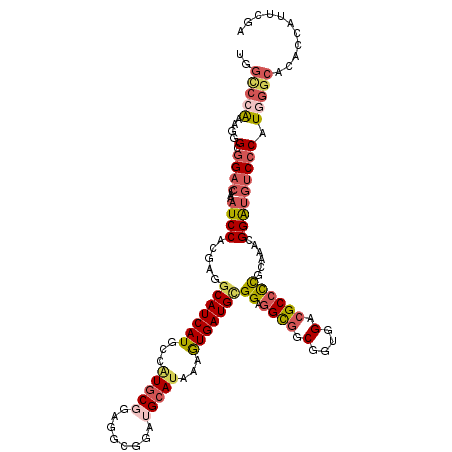
| Location | 7,344,022 – 7,344,142 |
| Length | 120 |
| Max. P | 0.543640 |

| Location | 7,344,022 – 7,344,142 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -52.23 |
| Consensus MFE | -28.50 |
| Energy contribution | -29.87 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
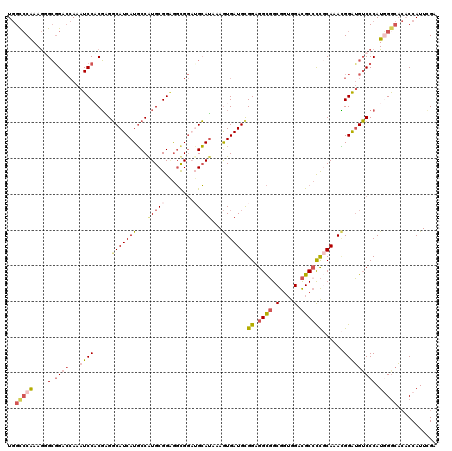
>2R_DroMel_CAF1 7344022 120 + 20766785 UGGCCCGAAGUGCGGACCAAAUCCACGAGGCAUCAUGCCAUGCGGAGGUGGAUGCAUAAAGUGAUGCGGAGGCGGCGGUGGGCGCCCCGCAAACGGAUGUCCCAUGGGCACACCAUUCGA ..(((((..(((((..(((..(((.((.(((.....))).)).)))..))).)))))...((..(((((.((((.(....).))))))))).))((.....)).)))))........... ( -50.80) >DroSec_CAF1 139628 120 + 1 UGGCCCGAAGUGCGGACCAAAUCCACGUGGCAUCAUGCCAUGCGGAGGUGGAUGCAUAAAGUGAUGCGGGGGCGGCGGUGGGCGCCCCGCAAACGGGUGUCCCAUGGACACACCAUUCGA .....(((((((((..(((..(((.((((((.....)))))).)))..))).)))))...((..(((((((.((.(....).))))))))).))((((((((...)))))).)).)))). ( -59.50) >DroEre_CAF1 139577 120 + 1 UGGCCCAAAGGGCGGACCAAAUCCACGGGGCAUCAUGCCGUGCGGAGGCGGAUGCAUAAAGUGAUGCGGAGGCGGCGGUGGACGCCCCGCAAACGGAUGUCCCAUGGGCACACCAUUCGA ..(((((..(((((..((.....(((...(((((.((((.......))))))))).....))).(((((.((((........)))))))))...)).)))))..)))))........... ( -53.00) >DroYak_CAF1 144104 120 + 1 UGGCCCAAAGGGCAGACCAAAUCCACGGGGCAUCAUGCCAUGCGGAGGUGGAUGCAUAAAGUGAUGCGGAGGCGGCGGUGGACGCCCCGCAAACGGAUGUCCCAUGGGCACACCAUUCGA ..(((((..(((((..(((..(((.((.(((.....))).)).)))..))).........((..(((((.((((........))))))))).))...)))))..)))))........... ( -51.50) >DroAna_CAF1 133209 120 + 1 UGGUGCAAAAGGAGGACCAAAUCCUCUCGUCAUCAUUCCAUGCGGCAUCGGUGGGAGAAAAUGAUGCGGUGGUGGCGGUGGACGCGCCCCAAAAGGAUGACCCAUUGGCACUCCAUUAGA .(((((....((((((.....)))))).((((((.......((.((((((...........)))))).))((.(((((....).)))))).....))))))......)))))........ ( -40.20) >DroPer_CAF1 105803 117 + 1 UAGCCCAAAGGGCGGAGGAAAUCCCCGAGGCAUCAUGCCGUGCGGGGGCG---GCAUGAAGUGAUGUGGCGGCUGCGCGGGAUGCCUUCCAAAUGGAAGUCCCAUCGGAUGCCCAUUGGG ...(((((.(((((((.....)))((((((((((...((((((((..((.---((((......)))).))..))))))))))))))((((....))))......))))..)))).))))) ( -58.40) >consensus UGGCCCAAAGGGCGGACCAAAUCCACGAGGCAUCAUGCCAUGCGGAGGCGGAUGCAUAAAGUGAUGCGGAGGCGGCGGUGGACGCCCCGCAAACGGAUGUCCCAUGGGCACACCAUUCGA ..(((((....(.((((...((((.....(((((((...(((((........)))))...)))))))((.((((.(....).))))))......))))))))).)))))........... (-28.50 = -29.87 + 1.37)

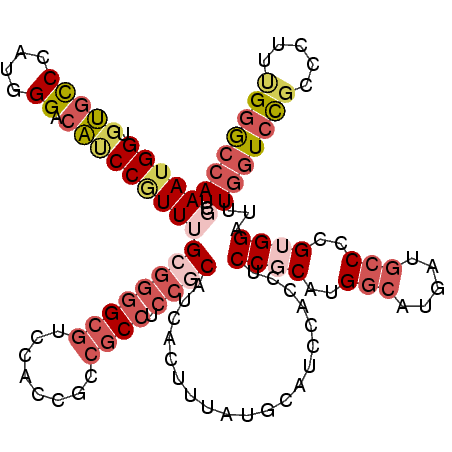
| Location | 7,344,022 – 7,344,142 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -48.15 |
| Consensus MFE | -26.96 |
| Energy contribution | -29.22 |
| Covariance contribution | 2.26 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
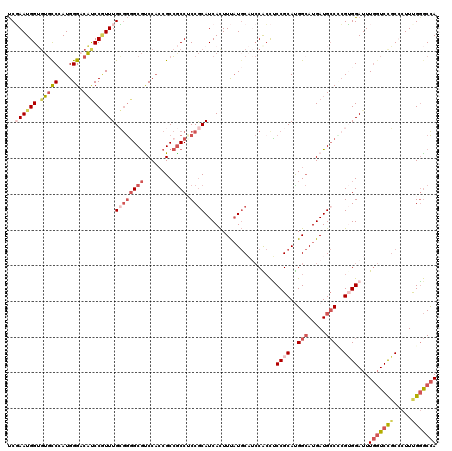
>2R_DroMel_CAF1 7344022 120 - 20766785 UCGAAUGGUGUGCCCAUGGGACAUCCGUUUGCGGGGCGCCCACCGCCGCCUCCGCAUCACUUUAUGCAUCCACCUCCGCAUGGCAUGAUGCCUCGUGGAUUUGGUCCGCACUUCGGGCCA ..(((((((((.((....))))).))))))((((((((........)))).))))...................(((((..(((.....)))..)))))..(((((((.....))))))) ( -46.80) >DroSec_CAF1 139628 120 - 1 UCGAAUGGUGUGUCCAUGGGACACCCGUUUGCGGGGCGCCCACCGCCGCCCCCGCAUCACUUUAUGCAUCCACCUCCGCAUGGCAUGAUGCCACGUGGAUUUGGUCCGCACUUCGGGCCA .(((((((.((((((...))))))))))))).((((((........)))))).((((......)))).......(((((.((((.....)))).)))))..(((((((.....))))))) ( -54.70) >DroEre_CAF1 139577 120 - 1 UCGAAUGGUGUGCCCAUGGGACAUCCGUUUGCGGGGCGUCCACCGCCGCCUCCGCAUCACUUUAUGCAUCCGCCUCCGCACGGCAUGAUGCCCCGUGGAUUUGGUCCGCCCUUUGGGCCA ........((.(((((.(((....(((((..(((((((((....((((.....((((......))))....((....)).))))..)))))))))..)))..))....)))..))))))) ( -51.30) >DroYak_CAF1 144104 120 - 1 UCGAAUGGUGUGCCCAUGGGACAUCCGUUUGCGGGGCGUCCACCGCCGCCUCCGCAUCACUUUAUGCAUCCACCUCCGCAUGGCAUGAUGCCCCGUGGAUUUGGUCUGCCCUUUGGGCCA ........((.(((((.(((....(((((..(((((((((....(((((....))........((((..........)))))))..)))))))))..)))..))....)))..))))))) ( -48.50) >DroAna_CAF1 133209 120 - 1 UCUAAUGGAGUGCCAAUGGGUCAUCCUUUUGGGGCGCGUCCACCGCCACCACCGCAUCAUUUUCUCCCACCGAUGCCGCAUGGAAUGAUGACGAGAGGAUUUGGUCCUCCUUUUGCACCA ......((((..((((......((((((((((((((.(....))))).))).((((((((....(((...((....))...))))))))).)))))))))))))..)))).......... ( -37.60) >DroPer_CAF1 105803 117 - 1 CCCAAUGGGCAUCCGAUGGGACUUCCAUUUGGAAGGCAUCCCGCGCAGCCGCCACAUCACUUCAUGC---CGCCCCCGCACGGCAUGAUGCCUCGGGGAUUUCCUCCGCCCUUUGGGCUA (((((.((((..((((.(((((((((....)))))...))))((((....)).........((((((---((........)))))))).)).))))(((.....))))))).)))))... ( -50.00) >consensus UCGAAUGGUGUGCCCAUGGGACAUCCGUUUGCGGGGCGUCCACCGCCGCCUCCGCAUCACUUUAUGCAUCCACCUCCGCAUGGCAUGAUGCCCCGUGGAUUUGGUCCGCCCUUUGGGCCA ..((((((.(((((....)).)))))))))((((((((........)))).))))....................((((..(((.....)))..))))...(((((((.....))))))) (-26.96 = -29.22 + 2.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:00 2006