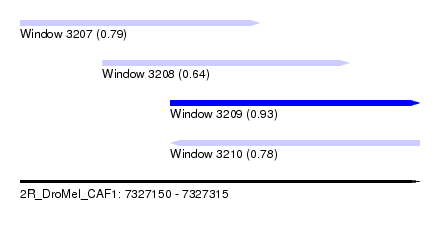
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,327,150 – 7,327,315 |
| Length | 165 |
| Max. P | 0.931693 |

| Location | 7,327,150 – 7,327,249 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -18.79 |
| Energy contribution | -20.57 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7327150 99 + 20766785 --CAGUAACAGCAGGGAAGAUCACCUGGACAAUUCCGACAUGUUGCUAGCGGGCGAGAACGAAGUGAAGCCACAACAGCCACGCAAGUUACUGUGGACAGU --(((((((((((((........)))(((....)))........))).(((((((....)...(((....)))....))).)))..)))))))........ ( -27.30) >DroPse_CAF1 99022 83 + 1 GGCGGUGGCAGCAAGGAGAACCACAGGCA------------GUGGCUGCAGGGCGACAAUAAGUUG------CAGCAGCAGCACAAGCUCCUGCUGCCACU ...((((((((((.((((...(((.....------------)))(((((...(((((.....))))------).)))))........)))))))))))))) ( -42.50) >DroSec_CAF1 122891 99 + 1 --CAGCAACAGCAGGGAAGAUCACCCGGAUAAUUCCGACAUGUUGCUGGCGGGCGAGAACGAAGUGAAGCCACAACAGCCACGCAAGUUGCUGUGGACAGU --((((((((.(.(((.......)))(((....))))...))))))))...(((..............)))...(((((.((....)).)))))....... ( -34.04) >DroSim_CAF1 127349 99 + 1 --CAGCAACAGCAGGGAAGAUCACCGGGAUAAUUCCGACAUGUUGCUGGCGGGCGAGAACGAAGUGAAGCCACUACAGCCACGCAAGUUGCUGUGGACAGU --((((((((...((((..(((.....)))..))))....))))))))...(((..............))).(((((((.((....)).)))))))..... ( -37.64) >DroEre_CAF1 122907 99 + 1 --CAGCCACAGCAGGGAAGAUCGCCCGGAUAAUUCGGACAUGUUGCUGGCGGGCGAGAACGAAGUGAAGCCACAACAGCCACGCAAGUUGCUGUGGACAGU --..((((((((((((.......))).........((...(((((.((((..((.........))...))))))))).))((....)))))))))).)... ( -34.90) >DroYak_CAF1 126719 99 + 1 --CAGCCACAGCAGGGAGGAUCGCCCGGAUAAUUCCGACAUGUUGCUGGCGGGCGAGAACGAAGUGAAGCCACAACUGCCACGCAAGUUGCUGUGGACAGU --..(((((((((((.((..(((((((((....))).....((.....)))))))).......(((....)))..)).))((....)))))))))).)... ( -39.20) >consensus __CAGCAACAGCAGGGAAGAUCACCCGGAUAAUUCCGACAUGUUGCUGGCGGGCGAGAACGAAGUGAAGCCACAACAGCCACGCAAGUUGCUGUGGACAGU ..((((.(((...((((..(((.....)))..))))....))).))))...(((..............)))...(((((.((....)).)))))....... (-18.79 = -20.57 + 1.78)


| Location | 7,327,184 – 7,327,286 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.99 |
| Mean single sequence MFE | -39.23 |
| Consensus MFE | -27.05 |
| Energy contribution | -29.67 |
| Covariance contribution | 2.61 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7327184 102 + 20766785 GACAUGUUGCUAGCGGGCGAGAACGAAGUGAAGCCACAACAGCCACGCAAGUUACUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACACUAGCA .......((((((((........))..(((..(((((.((((..((....))..))))(..(.(((....))).)..).......)))))..))).)))))) ( -30.70) >DroPse_CAF1 99051 82 + 1 -----GUGGCUGCAGGGCGACAAUAAGUUG------CAGCAGCAGCACAAGCUCCUGCUGCCACUGCAGCCGCCG------ACA---GGCAGCACGCGAGUA -----((((((((((((((((.....))).------.(((((.(((....))).)))))))).))))))))(((.------...---))).))((....)). ( -41.10) >DroSec_CAF1 122925 102 + 1 GACAUGUUGCUGGCGGGCGAGAACGAAGUGAAGCCACAACAGCCACGCAAGUUGCUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACGCUAGCA .......((((((((.((.........(((....)))....((((((...(((((.((((............))))..))))).)))))).)).)))))))) ( -39.70) >DroSim_CAF1 127383 102 + 1 GACAUGUUGCUGGCGGGCGAGAACGAAGUGAAGCCACUACAGCCACGCAAGUUGCUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACGCUAGCA .......((((((((.((......(.((((....)))).).((((((...(((((.((((............))))..))))).)))))).)).)))))))) ( -40.80) >DroEre_CAF1 122941 102 + 1 GACAUGUUGCUGGCGGGCGAGAACGAAGUGAAGCCACAACAGCCACGCAAGUUGCUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACGCCAGCA .......((((((((.((.........(((....)))....((((((...(((((.((((............))))..))))).)))))).)).)))))))) ( -42.00) >DroYak_CAF1 126753 102 + 1 GACAUGUUGCUGGCGGGCGAGAACGAAGUGAAGCCACAACUGCCACGCAAGUUGCUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACGCCUGCA ............(((((((........(((....)))..((((((((...(((((.((((............))))..))))).))))))))..))))))). ( -41.10) >consensus GACAUGUUGCUGGCGGGCGAGAACGAAGUGAAGCCACAACAGCCACGCAAGUUGCUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACGCUAGCA ........(((((((.((.........(((....)))....((((((...(((((.((((............))))..))))).)))))).)).))))))). (-27.05 = -29.67 + 2.61)

| Location | 7,327,212 – 7,327,315 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -26.81 |
| Energy contribution | -27.12 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7327212 103 + 20766785 UGAAGCCACAACAGCCACGCAAGUUACUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACACUAGCAACAUUGGUGGCAACACCAGAGCAGCAACA ....(((((.((((..((....))..))))(..(.(((....))).)..).......))))).........((..(.((((((....)))))).)..)).... ( -32.00) >DroPse_CAF1 99074 88 + 1 UG------CAGCAGCAGCACAAGCUCCUGCUGCCACUGCAGCCGCCG------ACA---GGCAGCACGCGAGUACCUUCCGCGACGACGCCCAGGAGGAGACU .(------((((((.(((....))).)))))))...(((.((.(((.------...---))).))..)))(((..((((((((....)))...)))))..))) ( -35.50) >DroSec_CAF1 122953 103 + 1 UGAAGCCACAACAGCCACGCAAGUUGCUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACGCUAGCAACAUUGGUGGCAACACCAGAGCAGCAACA ....(((((.(((((.((....)).)))))(..(.(((....))).)..).......))))).....(((.((....((((((....)))))).))))).... ( -38.30) >DroSim_CAF1 127411 103 + 1 UGAAGCCACUACAGCCACGCAAGUUGCUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACGCUAGCAACAUUGGUGGCAACACCAGAGCAGCAACA ...(((..(((((((.((....)).)))))))...((((..((((............))))..))))))).((..(.((((((....)))))).)..)).... ( -40.70) >DroEre_CAF1 122969 103 + 1 UGAAGCCACAACAGCCACGCAAGUUGCUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACGCCAGCAACACUGAUGGCGACACCAAUGCAGCAGCU ....(((((.(((((.((....)).)))))(..(.(((....))).)..).......))))).((((((((.((....)).))))).......)))....... ( -32.91) >DroYak_CAF1 126781 103 + 1 UGAAGCCACAACUGCCACGCAAGUUGCUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACGCCUGCAACAUUGGUGGCAACACCAGCGCAGCAACU ...........((((((((...(((((.((((............))))..))))).))))))))...((.(((....((((((....)))))).))))).... ( -39.10) >consensus UGAAGCCACAACAGCCACGCAAGUUGCUGUGGACAGUGCAACCACCGCUCGCAACACGUGGCAGCACGCUAGCAACAUUGGUGGCAACACCAGAGCAGCAACA ..........(((((.((....)).)))))((...((((..((((............))))..)))).)).((..(.((((((....)))))).)..)).... (-26.81 = -27.12 + 0.31)

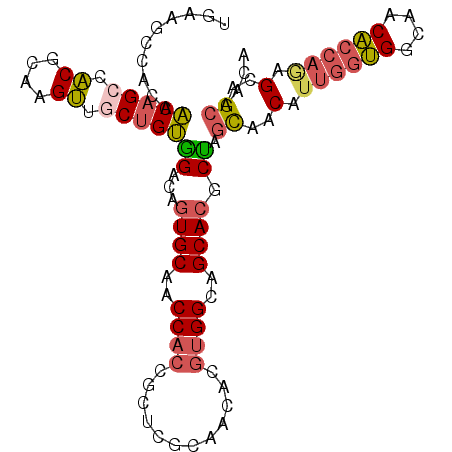
| Location | 7,327,212 – 7,327,315 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -42.04 |
| Consensus MFE | -28.86 |
| Energy contribution | -30.07 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7327212 103 - 20766785 UGUUGCUGCUCUGGUGUUGCCACCAAUGUUGCUAGUGUGCUGCCACGUGUUGCGAGCGGUGGUUGCACUGUCCACAGUAACUUGCGUGGCUGUUGUGGCUUCA ....((..(.........((((((((.((((((.((((((.(((((.(((.....)))))))).)))).....))))))))))).)))))....)..)).... ( -35.52) >DroPse_CAF1 99074 88 - 1 AGUCUCCUCCUGGGCGUCGUCGCGGAAGGUACUCGCGUGCUGCC---UGU------CGGCGGCUGCAGUGGCAGCAGGAGCUUGUGCUGCUGCUG------CA .((((......))))((((((((((..(((((....)))))..)---)).------))))))).((((..((((((.(....).))))))..)))------). ( -44.70) >DroSec_CAF1 122953 103 - 1 UGUUGCUGCUCUGGUGUUGCCACCAAUGUUGCUAGCGUGCUGCCACGUGUUGCGAGCGGUGGUUGCACUGUCCACAGCAACUUGCGUGGCUGUUGUGGCUUCA .(((((((....(((((.((((((....((((..(((((....)))))...))))..)))))).))))).....)))))))....(.((((.....)))).). ( -40.10) >DroSim_CAF1 127411 103 - 1 UGUUGCUGCUCUGGUGUUGCCACCAAUGUUGCUAGCGUGCUGCCACGUGUUGCGAGCGGUGGUUGCACUGUCCACAGCAACUUGCGUGGCUGUAGUGGCUUCA .((..((((...(((((.((((((....((((..(((((....)))))...))))..)))))).)))))(.((((.((.....))))))).))))..)).... ( -42.90) >DroEre_CAF1 122969 103 - 1 AGCUGCUGCAUUGGUGUCGCCAUCAGUGUUGCUGGCGUGCUGCCACGUGUUGCGAGCGGUGGUUGCACUGUCCACAGCAACUUGCGUGGCUGUUGUGGCUUCA .(((((.(((((((((....))))))))).)).)))((((.(((((.(((.....)))))))).)))).(.((((((((.((.....)).))))))))).... ( -43.70) >DroYak_CAF1 126781 103 - 1 AGUUGCUGCGCUGGUGUUGCCACCAAUGUUGCAGGCGUGCUGCCACGUGUUGCGAGCGGUGGUUGCACUGUCCACAGCAACUUGCGUGGCAGUUGUGGCUUCA ...(((.(((.(((((....))))).))).)))(((..(((((((((((((((..((((((....)))))).....)))))..))))))))))....)))... ( -45.30) >consensus AGUUGCUGCUCUGGUGUUGCCACCAAUGUUGCUAGCGUGCUGCCACGUGUUGCGAGCGGUGGUUGCACUGUCCACAGCAACUUGCGUGGCUGUUGUGGCUUCA ....(((((...(((((.((((((....((((..(((((....)))))...))))..)))))).)))))....(((((.((....)).))))).))))).... (-28.86 = -30.07 + 1.20)

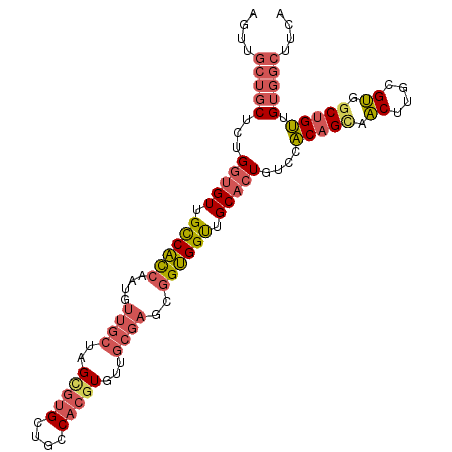

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:54 2006