
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,450,294 – 1,450,614 |
| Length | 320 |
| Max. P | 0.982435 |

| Location | 1,450,294 – 1,450,414 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -43.16 |
| Consensus MFE | -36.79 |
| Energy contribution | -37.80 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1450294 120 + 20766785 AUGAAGGUCAACGGCAGGCACGACUUUUUUUUGGCGUGUACUAUGCCAUUCCUUAGUGCUAUUGCCGUAGUUGUCGCGGCAGUUCAUAAAGCUCGCGACAACUGCAAGUGACUUGUGGAU ....((((((..(((((((((..........(((((((...))))))).......)))))..))))((((((((((((((..........)).))))))))))))...))))))...... ( -49.03) >DroSec_CAF1 68659 118 + 1 AUGAAGGUCAACGACAGGCACGACUUU--UUUGGCGUGUACUCCGCCAUUCCUUAGUGCUAUUAUCGUAGUUGUCGCGGCAGUUCAUAAAGCUCGCGACAGCUGCAAGUGACUUGUGGAU ....((((((......(((((......--..(((((.(....)))))).......)))))......((((((((((((((..........)).))))))))))))...))))))...... ( -43.86) >DroEre_CAF1 78289 119 + 1 AUGAAGGUCAACCGCAGGCAGGAUUC-UUUUUGGCAUGUACUACGCCACUCCUUGGUGCUAUUGUCGUAGUUGUCACAGCAGUUCAUAAAGCUCGCGACAGCUGCAAGUGACUUGUGGAU ...........(((((((((......-.....((((.((((((.(......).))))))...))))(((((((((.((((..........))).).)))))))))...)).))))))).. ( -36.60) >consensus AUGAAGGUCAACGGCAGGCACGACUUUUUUUUGGCGUGUACUACGCCAUUCCUUAGUGCUAUUGUCGUAGUUGUCGCGGCAGUUCAUAAAGCUCGCGACAGCUGCAAGUGACUUGUGGAU ....((((((..(((((((((..........((((((.....)))))).......)))))..))))((((((((((((((..........))).)))))))))))...))))))...... (-36.79 = -37.80 + 1.01)

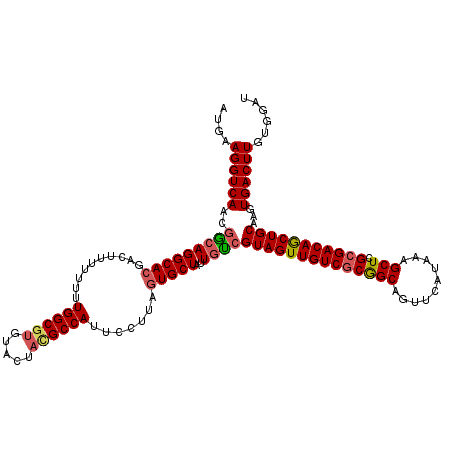
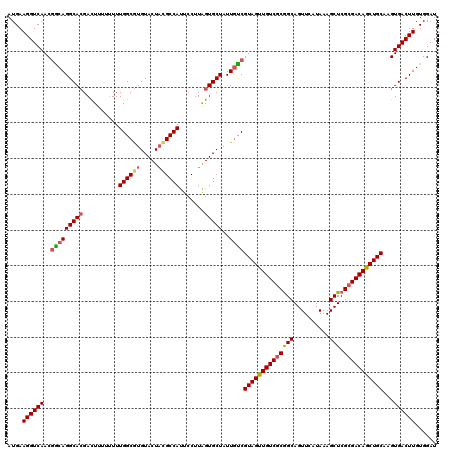
| Location | 1,450,334 – 1,450,454 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -46.03 |
| Consensus MFE | -35.27 |
| Energy contribution | -35.17 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1450334 120 + 20766785 CUAUGCCAUUCCUUAGUGCUAUUGCCGUAGUUGUCGCGGCAGUUCAUAAAGCUCGCGACAACUGCAAGUGACUUGUGGAUGCUCCUCCGGGGAAAAAGGAAUCGCGAGCGAGGAAUGGGA .....(((((((((.(((..((((((((.......)))))))).)))...((((((((((.(..((((...))))..).))((((...)))).........))))))))))))))))).. ( -50.20) >DroSec_CAF1 68697 120 + 1 CUCCGCCAUUCCUUAGUGCUAUUAUCGUAGUUGUCGCGGCAGUUCAUAAAGCUCGCGACAGCUGCAAGUGACUUGUGGAUGCUCCUCCGGCGAAGAAGGAGUCGCGAGCGAGGGACGGGA .((((...((((((...(((......((((((((((((((..........)).))))))))))))..(((((((.(...((((.....))))....).))))))).))))))))))))). ( -47.30) >DroEre_CAF1 78328 116 + 1 CUACGCCACUCCUUGGUGCUAUUGUCGUAGUUGUCACAGCAGUUCAUAAAGCUCGCGACAGCUGCAAGUGACUUGUGGAUGCUCCUCUGGGGAAGGAGGAAUCGCG----AGGAACGGGA ...(((((.....)))))........(((((((((.((((..........))).).)))))))))..((..(((((((.(.((((((....).))))).).)))))----))..)).... ( -40.60) >consensus CUACGCCAUUCCUUAGUGCUAUUGUCGUAGUUGUCGCGGCAGUUCAUAAAGCUCGCGACAGCUGCAAGUGACUUGUGGAUGCUCCUCCGGGGAAGAAGGAAUCGCGAGCGAGGAACGGGA .....((.((((((..(((.(((.((((((((((((((((..........))).)))))))))))......((.(.((.....)).).)).......))))).)))...)))))).)).. (-35.27 = -35.17 + -0.10)

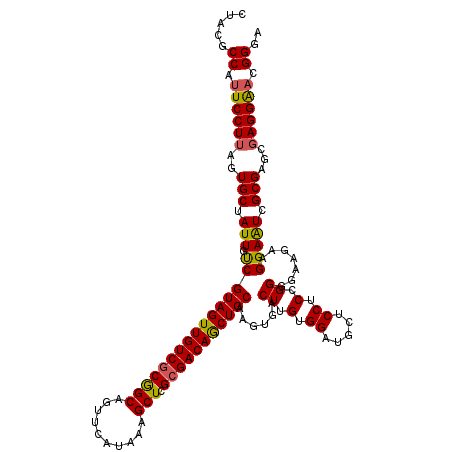
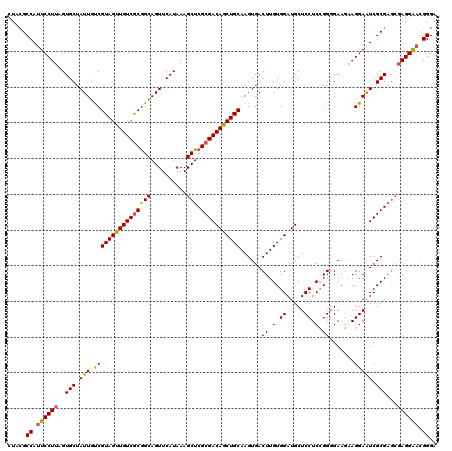
| Location | 1,450,374 – 1,450,494 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -41.97 |
| Consensus MFE | -31.01 |
| Energy contribution | -32.77 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1450374 120 + 20766785 AGUUCAUAAAGCUCGCGACAACUGCAAGUGACUUGUGGAUGCUCCUCCGGGGAAAAAGGAAUCGCGAGCGAGGAAUGGGAUGGUGCAGUUCGUUCGACUGCACUUCAUUAUUUUGCUCAU ..........((((((((((.(..((((...))))..).))((((...)))).........))))))))(((((((((...(((((((((.....)))))))))...))))))..))).. ( -42.70) >DroSec_CAF1 68737 120 + 1 AGUUCAUAAAGCUCGCGACAGCUGCAAGUGACUUGUGGAUGCUCCUCCGGCGAAGAAGGAGUCGCGAGCGAGGGACGGGAUGGUGCAGUUCGUUCGACUGCACUUCAUUAUUUUGCUCAU ..........(((((((((..(..((((...))))..)....((((.(......).)))))))))))))(((.((...((((((((((((.....))))))))..))))...)).))).. ( -47.00) >DroEre_CAF1 78368 106 + 1 AGUUCAUAAAGCUCGCGACAGCUGCAAGUGACUUGUGGAUGCUCCUCUGGGGAAGGAGGAAUCGCG----AGGAACGGGAUGGAGU----------GCUGCACUUCAUUAUUUUGCUCAU .((((......(((((((((.(..((((...))))..).)).((((((......)))))).)))))----))))))..((((((((----------(...)))))))))........... ( -36.20) >consensus AGUUCAUAAAGCUCGCGACAGCUGCAAGUGACUUGUGGAUGCUCCUCCGGGGAAGAAGGAAUCGCGAGCGAGGAACGGGAUGGUGCAGUUCGUUCGACUGCACUUCAUUAUUUUGCUCAU .((((.....((((((((((.(..((((...))))..).)).((((..........)))).))))))))...))))..((.(((((((((.....))))))))))).............. (-31.01 = -32.77 + 1.76)

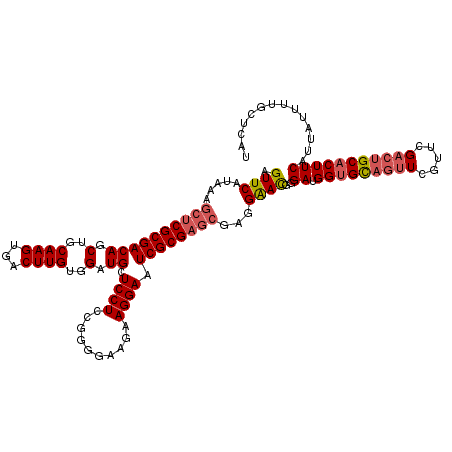
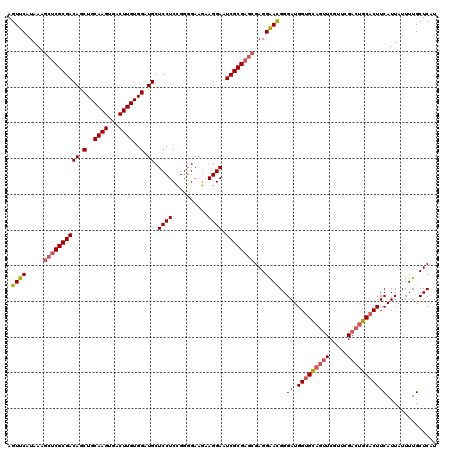
| Location | 1,450,454 – 1,450,574 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -21.19 |
| Energy contribution | -23.62 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1450454 120 + 20766785 UGGUGCAGUUCGUUCGACUGCACUUCAUUAUUUUGCUCAUUUCCUUUUAACGACAUUAUCAAUGGCUAUUUUACGCCAUUGUUUCGCAUUGCAUAGUCUAUUGUCCCAGCUGGGAUGUCA .(((((((((.....)))))))))...........................(((.....(((((((........))))))).....................(((((....)))))))). ( -31.90) >DroSec_CAF1 68817 120 + 1 UGGUGCAGUUCGUUCGACUGCACUUCAUUAUUUUGCUCAUUUCCUCUUAACGACAUUCUCAAUGGCUAUUUUGCGCCAUUGUUUCGCAUUGCAUAGUCUAUUGUCCCAGCUGGGAUGUCA .(((((((((.....)))))))))...........................(((((((((((((((........)))))))....((...(((........)))....)).)))))))). ( -32.70) >DroEre_CAF1 78444 110 + 1 UGGAGU----------GCUGCACUUCAUUAUUUUGCUCAUUUCCUUUUAACGACAUUAUCAAUGGCUAUUUUACGCCAUUGUUUCGCAUUGCAUAGUCUAUUGUCCCAGCUGGGAUGUCA ((((((----------(...)))))))........................(((.....(((((((........))))))).....................(((((....)))))))). ( -25.80) >DroYak_CAF1 73062 105 + 1 -----U----------GCUGCACUUCAUUAUUUUGCUCAUUUCCUUUUAACGACAUUAUCAAUGGUUAUUUUACGCCAUUGUUUCGCAUUGAAUAGUCUAUUGUCCCAGCUGGGAUGUCA -----.----------...(((...........)))...............(((.....(((((((........))))))).....................(((((....)))))))). ( -18.20) >consensus UGGUGC__________ACUGCACUUCAUUAUUUUGCUCAUUUCCUUUUAACGACAUUAUCAAUGGCUAUUUUACGCCAUUGUUUCGCAUUGCAUAGUCUAUUGUCCCAGCUGGGAUGUCA .(((((((((.....)))))))))...........................(((.....(((((((........))))))).....................(((((....)))))))). (-21.19 = -23.62 + 2.44)

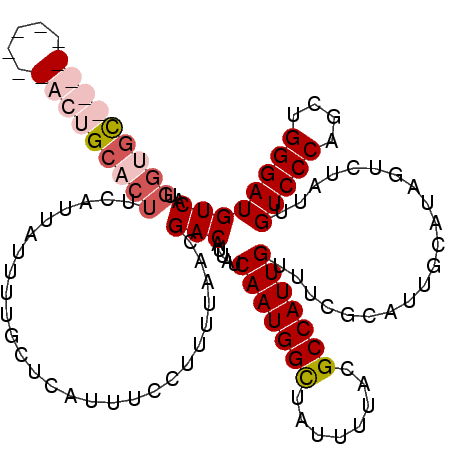
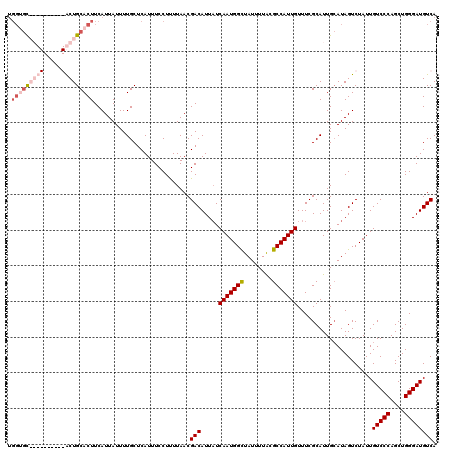
| Location | 1,450,454 – 1,450,574 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -20.25 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1450454 120 - 20766785 UGACAUCCCAGCUGGGACAAUAGACUAUGCAAUGCGAAACAAUGGCGUAAAAUAGCCAUUGAUAAUGUCGUUAAAAGGAAAUGAGCAAAAUAAUGAAGUGCAGUCGAACGAACUGCACCA ((((.((((....)))).....(((..(((...)))...(((((((........))))))).....)))))))........................((((((((....).))))))).. ( -31.70) >DroSec_CAF1 68817 120 - 1 UGACAUCCCAGCUGGGACAAUAGACUAUGCAAUGCGAAACAAUGGCGCAAAAUAGCCAUUGAGAAUGUCGUUAAGAGGAAAUGAGCAAAAUAAUGAAGUGCAGUCGAACGAACUGCACCA ((((.((((....)))).....(((..(((...)))...(((((((........))))))).....)))))))........................((((((((....).))))))).. ( -31.70) >DroEre_CAF1 78444 110 - 1 UGACAUCCCAGCUGGGACAAUAGACUAUGCAAUGCGAAACAAUGGCGUAAAAUAGCCAUUGAUAAUGUCGUUAAAAGGAAAUGAGCAAAAUAAUGAAGUGCAGC----------ACUCCA ((((.((((....)))).....(((..(((...)))...(((((((........))))))).....)))))))...(((.....(((...........)))...----------..))). ( -25.20) >DroYak_CAF1 73062 105 - 1 UGACAUCCCAGCUGGGACAAUAGACUAUUCAAUGCGAAACAAUGGCGUAAAAUAACCAUUGAUAAUGUCGUUAAAAGGAAAUGAGCAAAAUAAUGAAGUGCAGC----------A----- ((((.((((....)))).....(((..(((.....))).((((((..........)))))).....)))))))...........(((...........)))...----------.----- ( -17.40) >consensus UGACAUCCCAGCUGGGACAAUAGACUAUGCAAUGCGAAACAAUGGCGUAAAAUAGCCAUUGAUAAUGUCGUUAAAAGGAAAUGAGCAAAAUAAUGAAGUGCAGC__________ACACCA .((((((((....))))......................(((((((........)))))))....))))...............(((...........)))................... (-20.25 = -20.50 + 0.25)

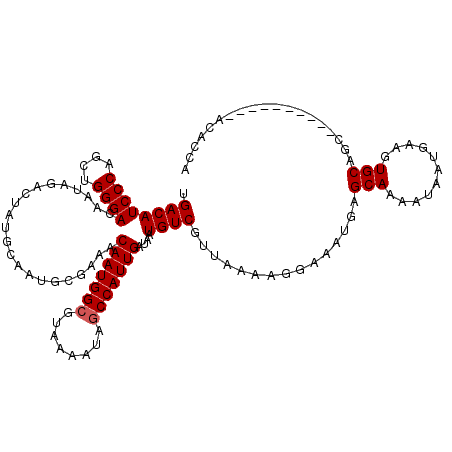
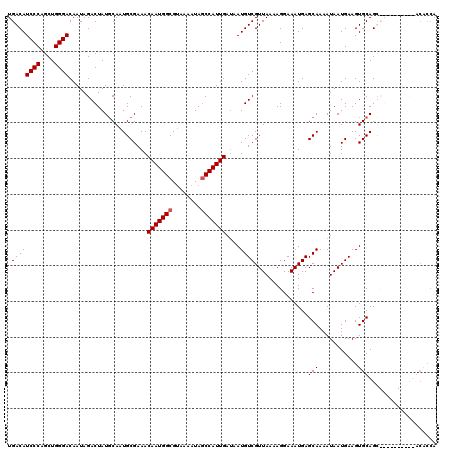
| Location | 1,450,494 – 1,450,614 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -22.09 |
| Energy contribution | -22.15 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1450494 120 + 20766785 UUCCUUUUAACGACAUUAUCAAUGGCUAUUUUACGCCAUUGUUUCGCAUUGCAUAGUCUAUUGUCCCAGCUGGGAUGUCAUUUUAUUUGAUUUUCAAUUACGUUUAAUUAAUUAAGUGUA ...........(((.....(((((((........))))))).....................(((((....))))))))....(((((((((...(((((....)))))))))))))).. ( -23.20) >DroSec_CAF1 68857 120 + 1 UUCCUCUUAACGACAUUCUCAAUGGCUAUUUUGCGCCAUUGUUUCGCAUUGCAUAGUCUAUUGUCCCAGCUGGGAUGUCAUUUUAUUUGAUUUUCAAUUACGUUUAAUUAAUUAAGUGUA ...........(((((((((((((((........)))))))....((...(((........)))....)).))))))))....(((((((((...(((((....)))))))))))))).. ( -24.00) >DroEre_CAF1 78474 120 + 1 UUCCUUUUAACGACAUUAUCAAUGGCUAUUUUACGCCAUUGUUUCGCAUUGCAUAGUCUAUUGUCCCAGCUGGGAUGUCAUUUUAUUUGAUUUUCAAUUACGUUUAAUUAAUUAAGUGUA ...........(((.....(((((((........))))))).....................(((((....))))))))....(((((((((...(((((....)))))))))))))).. ( -23.20) >DroYak_CAF1 73087 120 + 1 UUCCUUUUAACGACAUUAUCAAUGGUUAUUUUACGCCAUUGUUUCGCAUUGAAUAGUCUAUUGUCCCAGCUGGGAUGUCAUUUUAUUUGAUUUUCAAUUACUCUUAAUUAAUUAAUUGUA ..........(((......(((((((........)))))))..))).((..(((((.....((((((....)))))).....)))))..))...((((((............)))))).. ( -20.00) >consensus UUCCUUUUAACGACAUUAUCAAUGGCUAUUUUACGCCAUUGUUUCGCAUUGCAUAGUCUAUUGUCCCAGCUGGGAUGUCAUUUUAUUUGAUUUUCAAUUACGUUUAAUUAAUUAAGUGUA ...........(((.....(((((((........))))))).....................(((((....))))))))....(((((((((...(((((....)))))))))))))).. (-22.09 = -22.15 + 0.06)

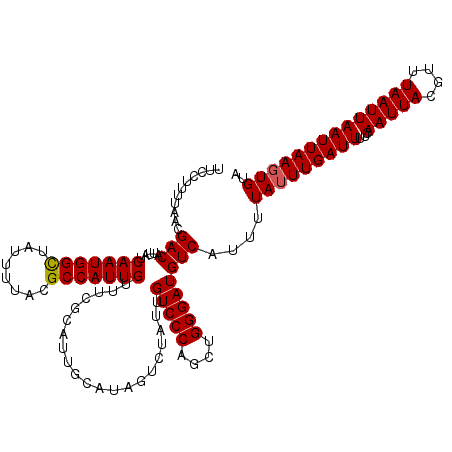
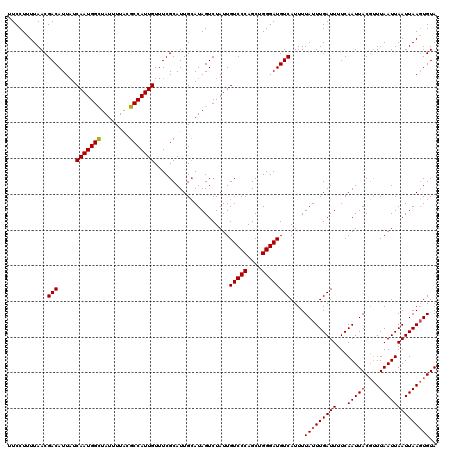
| Location | 1,450,494 – 1,450,614 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -23.45 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1450494 120 - 20766785 UACACUUAAUUAAUUAAACGUAAUUGAAAAUCAAAUAAAAUGACAUCCCAGCUGGGACAAUAGACUAUGCAAUGCGAAACAAUGGCGUAAAAUAGCCAUUGAUAAUGUCGUUAAAAGGAA ....(((..(((((((....)))))))...........(((((((((((....))))......................(((((((........)))))))....)))))))..)))... ( -25.80) >DroSec_CAF1 68857 120 - 1 UACACUUAAUUAAUUAAACGUAAUUGAAAAUCAAAUAAAAUGACAUCCCAGCUGGGACAAUAGACUAUGCAAUGCGAAACAAUGGCGCAAAAUAGCCAUUGAGAAUGUCGUUAAGAGGAA ....(((..(((((((....)))))))...........(((((((((((....))))......................(((((((........)))))))....)))))))..)))... ( -26.20) >DroEre_CAF1 78474 120 - 1 UACACUUAAUUAAUUAAACGUAAUUGAAAAUCAAAUAAAAUGACAUCCCAGCUGGGACAAUAGACUAUGCAAUGCGAAACAAUGGCGUAAAAUAGCCAUUGAUAAUGUCGUUAAAAGGAA ....(((..(((((((....)))))))...........(((((((((((....))))......................(((((((........)))))))....)))))))..)))... ( -25.80) >DroYak_CAF1 73087 120 - 1 UACAAUUAAUUAAUUAAGAGUAAUUGAAAAUCAAAUAAAAUGACAUCCCAGCUGGGACAAUAGACUAUUCAAUGCGAAACAAUGGCGUAAAAUAACCAUUGAUAAUGUCGUUAAAAGGAA ..((((((.((......)).))))))............(((((((((((....))))......................((((((..........))))))....)))))))........ ( -20.50) >consensus UACACUUAAUUAAUUAAACGUAAUUGAAAAUCAAAUAAAAUGACAUCCCAGCUGGGACAAUAGACUAUGCAAUGCGAAACAAUGGCGUAAAAUAGCCAUUGAUAAUGUCGUUAAAAGGAA .........(((((((....)))))))...........(((((((((((....))))......................(((((((........)))))))....)))))))........ (-23.45 = -23.70 + 0.25)

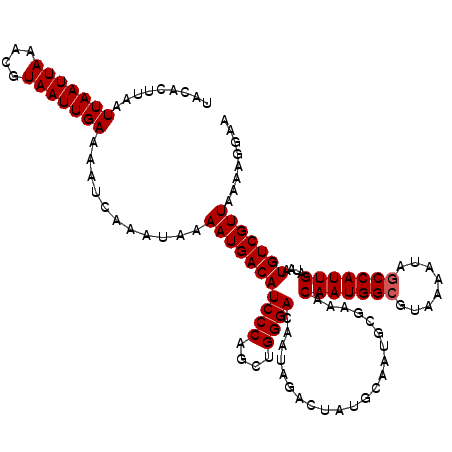
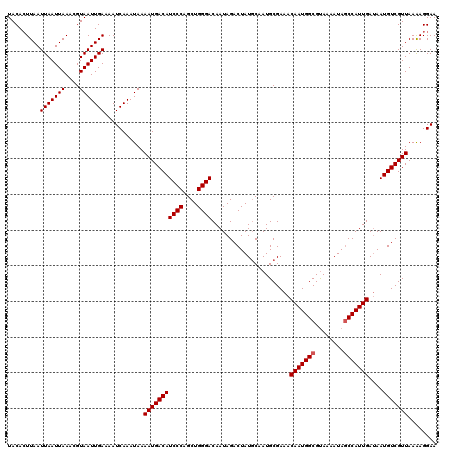
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:06 2006