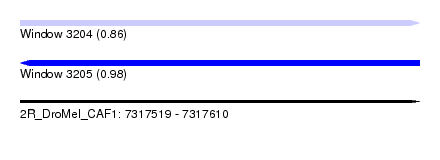
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,317,519 – 7,317,610 |
| Length | 91 |
| Max. P | 0.980420 |

| Location | 7,317,519 – 7,317,610 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7317519 91 + 20766785 UGUUGCAACUGCAAGCUGCAACUG-------CAAAUGCCACAGGCGAGUGUUAACGUAUACCCAUUAAAUGGGAUACCGAAAGGCGUAAACAAGUGAC ..(((((..(((.....)))..))-------))).((((...))))..((((.(((....(((((...)))))...((....))))).))))...... ( -25.00) >DroSec_CAF1 113098 91 + 1 UGUUGCAACUGCAAGCUGCAACUG-------CAAAUGCCACAGACGAGUGUUAACGUAUACCCAUUAAAUGGGAUACCGAAAGGCGUAAACAAGUGAC ..(((((..(((.....)))..))-------)))..........((..((((.(((....(((((...)))))...((....))))).))))..)).. ( -24.20) >DroSim_CAF1 117561 91 + 1 UGUUGCAACUGCAAGCUGCAACUG-------CAAAUGCCACAGUCGAGUGUUAACGUAUACCCAUUAAAUGGGAUACCGAAAGGCGUAAACAAGUGAC ..(((((..(((.....)))..))-------)))........((((..((((.(((....(((((...)))))...((....))))).))))..)))) ( -27.40) >DroEre_CAF1 113141 98 + 1 UGUUGCAACUGCAAGCUGCAACUGCAAGCUGCAACUGCCGCAGAGGAGUGUUAACGUAUACCCAUUAAAUGGUAUACCGAAAGGCGUAAACAAGUGAC .((..(..((((.((((((....)).))))((....)).)))).....((((.(((((((((........))))))((....))))).)))).)..)) ( -29.60) >DroYak_CAF1 116089 91 + 1 UGUUGCAACUGCAAGCUGCAACUG-------CAACUGCCUCAGUCGAGUGUUAACGUAUACCCAUUAAAUGGGAUACCGAAAGGCGUAAACAAGUGAC .((((((..(((.....)))..))-------)))).......((((..((((.(((....(((((...)))))...((....))))).))))..)))) ( -30.30) >consensus UGUUGCAACUGCAAGCUGCAACUG_______CAAAUGCCACAGACGAGUGUUAACGUAUACCCAUUAAAUGGGAUACCGAAAGGCGUAAACAAGUGAC .((((((.(.....).))))))......................((..((((.(((....(((((...)))))...((....))))).))))..)).. (-20.50 = -20.90 + 0.40)

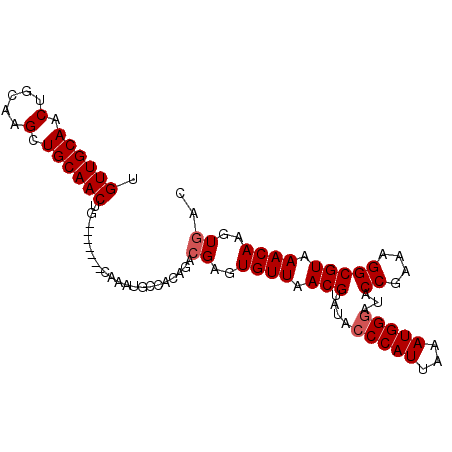
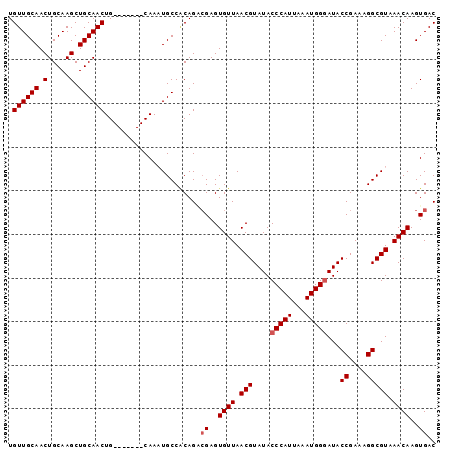
| Location | 7,317,519 – 7,317,610 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -22.68 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7317519 91 - 20766785 GUCACUUGUUUACGCCUUUCGGUAUCCCAUUUAAUGGGUAUACGUUAACACUCGCCUGUGGCAUUUG-------CAGUUGCAGCUUGCAGUUGCAACA (((((..((...((.....))(((((((((...))))).))))..........))..))))).....-------..((((((((.....)))))))). ( -26.70) >DroSec_CAF1 113098 91 - 1 GUCACUUGUUUACGCCUUUCGGUAUCCCAUUUAAUGGGUAUACGUUAACACUCGUCUGUGGCAUUUG-------CAGUUGCAGCUUGCAGUUGCAACA ......(((..(.(((.....(((((((((...))))).))))....(((......)))))).)..)-------))((((((((.....)))))))). ( -26.10) >DroSim_CAF1 117561 91 - 1 GUCACUUGUUUACGCCUUUCGGUAUCCCAUUUAAUGGGUAUACGUUAACACUCGACUGUGGCAUUUG-------CAGUUGCAGCUUGCAGUUGCAACA (((((..(((..((.....))(((((((((...))))).))))..........))).))))).....-------..((((((((.....)))))))). ( -27.00) >DroEre_CAF1 113141 98 - 1 GUCACUUGUUUACGCCUUUCGGUAUACCAUUUAAUGGGUAUACGUUAACACUCCUCUGCGGCAGUUGCAGCUUGCAGUUGCAGCUUGCAGUUGCAACA .............(((.....(((((((........)))))))((............))))).(((((((((.(((((....)).)))))))))))). ( -30.50) >DroYak_CAF1 116089 91 - 1 GUCACUUGUUUACGCCUUUCGGUAUCCCAUUUAAUGGGUAUACGUUAACACUCGACUGAGGCAGUUG-------CAGUUGCAGCUUGCAGUUGCAACA .............(((((((((((((((((...))))).))))(....)...)))..))))).((((-------((..(((.....)))..)))))). ( -31.10) >consensus GUCACUUGUUUACGCCUUUCGGUAUCCCAUUUAAUGGGUAUACGUUAACACUCGACUGUGGCAUUUG_______CAGUUGCAGCUUGCAGUUGCAACA (((((.((((.(((((....)))..(((((...))))).....)).)))).......)))))..............((((((((.....)))))))). (-22.68 = -23.28 + 0.60)

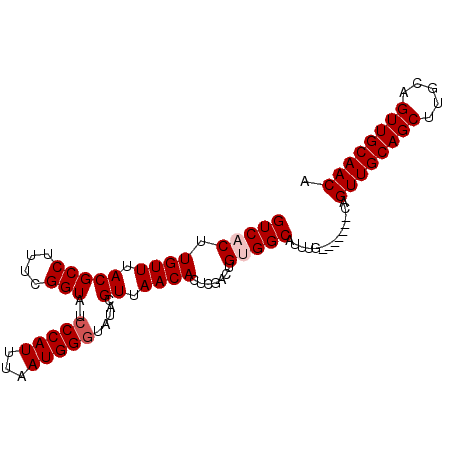
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:49 2006