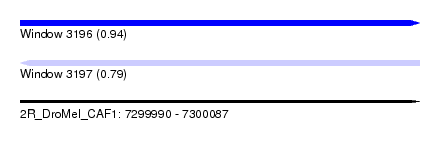
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,299,990 – 7,300,087 |
| Length | 97 |
| Max. P | 0.940128 |

| Location | 7,299,990 – 7,300,087 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 113 |
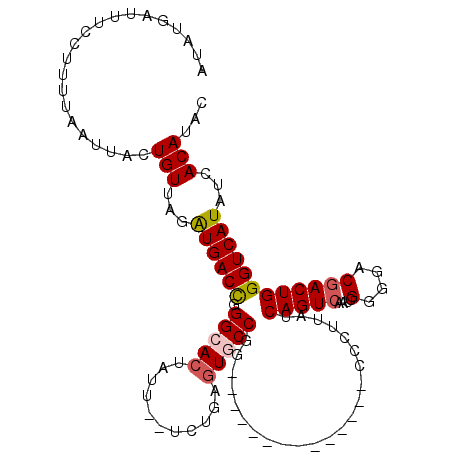
| Reading direction | forward |
| Mean pairwise identity | 80.96 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -21.09 |
| Energy contribution | -21.05 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7299990 97 + 20766785 GUAUGUGAUAUGACCCAGUCGUCCCCGAUUGACUGAUAAGGC--------------CCGGCACUCAGA--AAUAGUGCCCGGUCACUUAACAGUAAUUAAAGGUAAAUCAUAU ..(((((((...(((((((((....))))))((((.((((((--------------(.((((((.(..--..))))))).)))..)))).)))).......)))..))))))) ( -34.20) >DroSec_CAF1 95712 97 + 1 GUAUGUGAUAUGACCCAGUCGUCCCCGAUUGACUGAUAAGGU--------------CCGGCACUCAGA--AAUUGUGCCCGGUCAUCUAACAGUAAUUAAAAGGAAAUCAUAU ..(((((((.(((((((((((........)))))).......--------------..(((((.....--....))))).)))))(((..............))).))))))) ( -25.14) >DroSim_CAF1 99686 97 + 1 GUAUGUGAUAUGACCCAGUCGUCCCCGAUUGACUGAUAAGGC--------------CCGGUACUCAGA--AUAAGUGCCCGGUCAUCUAACAGUAAUUAAAAGGAAAUCAUAU ..(((((((....((((((((....))))))((((.((.(((--------------(.((((((....--...)))))).))))...)).))))........))..))))))) ( -26.60) >DroEre_CAF1 96023 109 + 1 GUAUGUGAUAUGACCCAGUUGUCC-CGAUUGACUGAUAAGCGUGGUUUCUGGUAGUCUGGUACUC---AGAAUACUGCCCGGUCAUCAAACACUAAUUAAUAAGGAAUCACAC ...((((((((((((((((((...-....))))))....(.(..((((((((((......))).)---)))).))..).)))))))......((........))..)))))). ( -26.70) >DroYak_CAF1 98178 99 + 1 GUAUGUGAUAUGACCCAGUCGUCCCCGCUUGACUGAUAAGAG--------------CUGGCACUCGGCACAAUUGUGCCCAGUCAUCAAACAGUAAUUAAAAGGGAAUCACAU ..((((((((((((...))))).(((...((((((....(((--------------......)))(((((....))))))))))).................))).))))))) ( -29.05) >consensus GUAUGUGAUAUGACCCAGUCGUCCCCGAUUGACUGAUAAGGC______________CCGGCACUCAGA__AAUAGUGCCCGGUCAUCUAACAGUAAUUAAAAGGAAAUCAUAU ...((((((((((((((((((........)))))).......................(((((...........))))).))))))....................)))))). (-21.09 = -21.05 + -0.04)

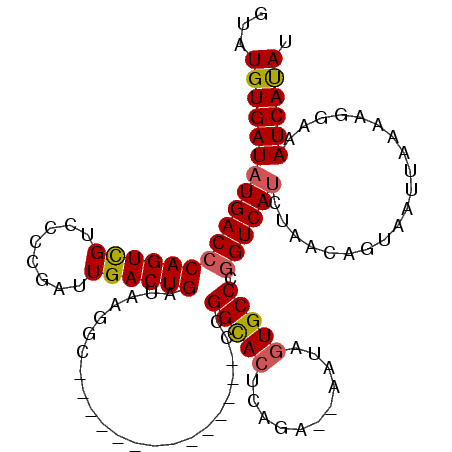
| Location | 7,299,990 – 7,300,087 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.96 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -18.80 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7299990 97 - 20766785 AUAUGAUUUACCUUUAAUUACUGUUAAGUGACCGGGCACUAUU--UCUGAGUGCCGG--------------GCCUUAUCAGUCAAUCGGGGACGACUGGGUCAUAUCACAUAC ((((((((((.........((((.((((.(.((.((((((...--....)))))).)--------------)))))).))))...(((....))).))))))))))....... ( -32.50) >DroSec_CAF1 95712 97 - 1 AUAUGAUUUCCUUUUAAUUACUGUUAGAUGACCGGGCACAAUU--UCUGAGUGCCGG--------------ACCUUAUCAGUCAAUCGGGGACGACUGGGUCAUAUCACAUAC .((((........(((((....)))))((((((.(((((....--.....)))))..--------------...........((.(((....))).))))))))....)))). ( -24.70) >DroSim_CAF1 99686 97 - 1 AUAUGAUUUCCUUUUAAUUACUGUUAGAUGACCGGGCACUUAU--UCUGAGUACCGG--------------GCCUUAUCAGUCAAUCGGGGACGACUGGGUCAUAUCACAUAC ((((((((...........((((.(((....((((..((((..--...)))).))))--------------...))).))))((.(((....))).))))))))))....... ( -24.20) >DroEre_CAF1 96023 109 - 1 GUGUGAUUCCUUAUUAAUUAGUGUUUGAUGACCGGGCAGUAUUCU---GAGUACCAGACUACCAGAAACCACGCUUAUCAGUCAAUCG-GGACAACUGGGUCAUAUCACAUAC (((((((.((...........((.(((((((.((((.....((((---(.(((......)))))))).)).)).))))))).))..((-(.....)))))....))))))).. ( -25.20) >DroYak_CAF1 98178 99 - 1 AUGUGAUUCCCUUUUAAUUACUGUUUGAUGACUGGGCACAAUUGUGCCGAGUGCCAG--------------CUCUUAUCAGUCAAGCGGGGACGACUGGGUCAUAUCACAUAC (((((((.(((.................(((((((((((....)))))((((....)--------------)))....))))))((((....)).)))))....))))))).. ( -37.10) >consensus AUAUGAUUUCCUUUUAAUUACUGUUAGAUGACCGGGCACUAUU__UCUGAGUGCCGG______________CCCUUAUCAGUCAAUCGGGGACGACUGGGUCAUAUCACAUAC .....................(((...((((((.(((((...........))))).......................(((((....(....))))))))))))...)))... (-18.80 = -19.28 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:42 2006