
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,298,572 – 7,298,692 |
| Length | 120 |
| Max. P | 0.994597 |

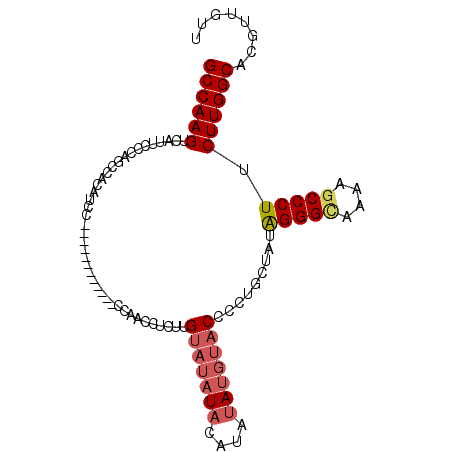
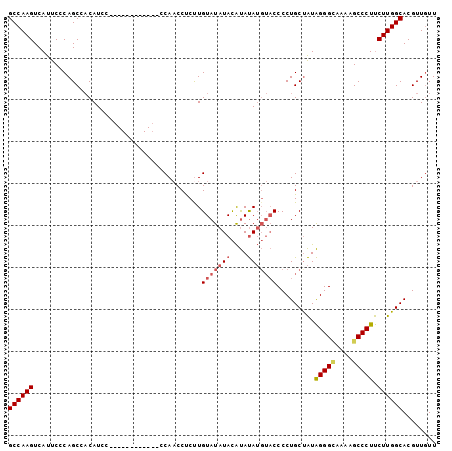
| Location | 7,298,572 – 7,298,662 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -16.99 |
| Energy contribution | -18.18 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7298572 90 + 20766785 GCCAAGUCAUUCCCAGCCACAUCC------------CCAACCUCUUGUAUAUACAUAUAUGUACCCCUGCCAUAGGGUAAAAGCCCUUCUUGGCACGUUGUU ........................------------.((((.....(((((((....)))))))...(((((.(((((....)))))...))))).)))).. ( -20.60) >DroSec_CAF1 94288 90 + 1 GCCAAGUCAUUCCCAGCCACAUCC------------CCAACCUCUUGUAUAUACAUAUAUGUACCCCUGCUAUAGGGCGAAAGCCCUUCUUGGCACGUUGUU ((((((..................------------..........(((((((....))))))).........(((((....))))).))))))........ ( -24.70) >DroSim_CAF1 98254 90 + 1 GCCAAGUCAUUCCCAGCCACAUCC------------CCAACCCCCUGUAUAUACAUAUAUGUACCCCUGCUAUAGGGCAAAAGCCCUUCUUGGCACGUUGUU ((((((..................------------..........(((((((....))))))).........(((((....))))).))))))........ ( -21.00) >DroEre_CAF1 94644 98 + 1 GCCAAGUCAUCCGCAGCCCCAUCCCCAUCCCCAUCCCCAUCCUCUUGUAUAUAUGCACA---CCCCCUGCUGUGGGGAA-AAGCCCUCCUUGGCACGUUAUU ((((((......((..((((((...............((......)).......(((..---.....))).))))))..-..))....))))))........ ( -19.80) >consensus GCCAAGUCAUUCCCAGCCACAUCC____________CCAACCUCUUGUAUAUACAUAUAUGUACCCCUGCUAUAGGGCAAAAGCCCUUCUUGGCACGUUGUU ((((((........................................(((((((....))))))).........(((((....))))).))))))........ (-16.99 = -18.18 + 1.19)

| Location | 7,298,596 – 7,298,692 |
|---|---|
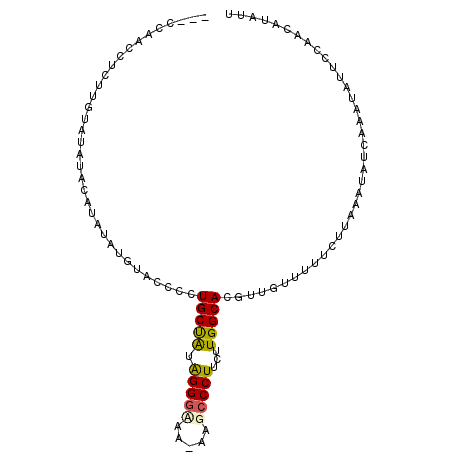
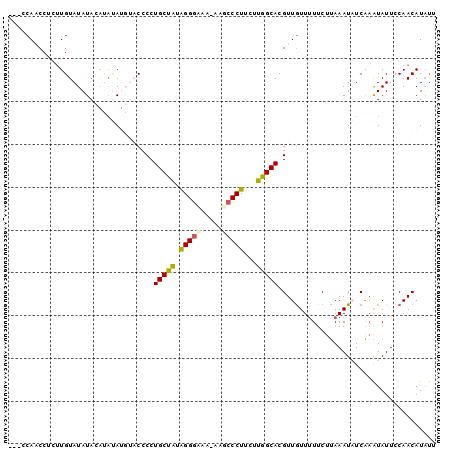
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.08 |
| Mean single sequence MFE | -18.15 |
| Consensus MFE | -11.02 |
| Energy contribution | -11.06 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7298596 96 + 20766785 ---CCAACCUCUUGUAUAUACAUAUAUGUACCCCUGCCAUAGGGUAA-AAGCCCUUCUUGGCACGUUGUUUUUCUUAAAUAUCAAAUAUUCCAACAUAUU ---.((((.....(((((((....)))))))...(((((.(((((..-..)))))...))))).))))................................ ( -20.60) >DroSec_CAF1 94312 93 + 1 ---CCAACCUCUUGUAUAUACAUAUAUGUACCCCUGCUAUAGGGCGA-AAGCCCUUCUUGGCACGUUGUUUUUCUUAAAUAUCAAAUAUUCCAACAU--- ---.((((.....(((((((....)))))))...(((((.(((((..-..)))))...))))).)))).............................--- ( -23.90) >DroSim_CAF1 98278 96 + 1 ---CCAACCCCCUGUAUAUACAUAUAUGUACCCCUGCUAUAGGGCAA-AAGCCCUUCUUGGCACGUUGUUUUUCUUAAAUAUCAAAUAUUCCAACAUAUU ---.((((.....(((((((....)))))))...(((((.(((((..-..)))))...))))).))))................................ ( -20.20) >DroEre_CAF1 94677 95 + 1 UCCCCAUCCUCUUGUAUAUAUGCACA---CCCCCUGCUGUGGGGAA--AAGCCCUCCUUGGCACGUUAUUUUUCUCAAAUUUCGAGUAUUCCAACAUAUU (((((((..............(((..---.....))).))))))).--..(((......)))..(((......(((.......)))......)))..... ( -16.46) >DroYak_CAF1 96758 92 + 1 UCCCCAUCCUCUUGUAUAUA----GA---CCCCCUGCUAUAGGAAAAAAACCCCUCCUUGGCACGUUGUU-UUCUUAAAUUUCAUAUAUUCCAACAUAUU .............((((((.----..---.....(((((.((((..........))))))))).(..(((-(....))))..)))))))........... ( -9.60) >consensus ___CCAACCUCUUGUAUAUACAUAUAUGUACCCCUGCUAUAGGGAAA_AAGCCCUUCUUGGCACGUUGUUUUUCUUAAAUAUCAAAUAUUCCAACAUAUU ..................................(((((.(((((.....)))))...)))))..................................... (-11.02 = -11.06 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:40 2006