
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
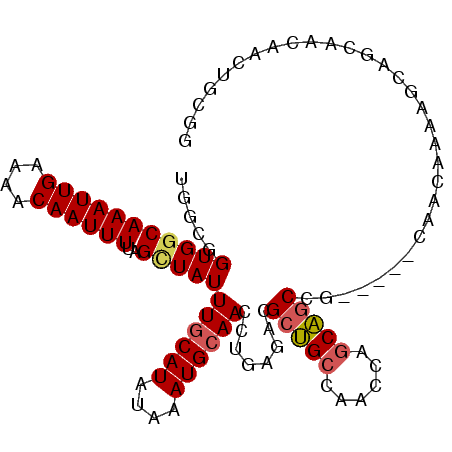
| Location | 7,282,440 – 7,282,539 |
| Length | 99 |
| Max. P | 0.799603 |

| Location | 7,282,440 – 7,282,539 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -14.92 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7282440 99 + 20766785 UGGCGGUGGCAAAUUGAAAACAAUUUUAGCUAUUUGCAUAUAAAUGCAACUUGAGACGCUGCCAACCAGCAGCCG-----CAACAAAAGCAGCAACAACUGCUG ..((((((((((((((....))))))..)))))((((((....))))))........(((((......)))))))-----)......(((((......))))). ( -28.40) >DroPse_CAF1 66332 99 + 1 GGACAGUGGCAAAUUGAAAACAAUUUUAGUUAUUUGCAUAUAAAUGCAACUUGAGACGGCGGCGGCAGGCGGCAC-----GGGGUGGAGGAGGCUCUAAGGGAU ....((((((((((((....))))))..))))))(((((....))))).(((....((((.((.....)).)).)-----)...(((((....))))))))... ( -20.30) >DroSec_CAF1 78340 99 + 1 UGGCGGUGGCAAAUUGAAAACAAUUUUAGCUAUUUGCAUAUAAAUGCAACCUGAGACGCUGCCAACCAGCAGCCG-----CAACAAAAGCAGCAACAACUGCGG ((((((((((((((((....))))))..))...((((((....)))))).......)))))))).((.((....)-----).......((((......)))))) ( -28.70) >DroSim_CAF1 81815 99 + 1 UGGCGGUGGCAAAUUGAAAACAAUUUUAGCUAUUUGCAUAUAAAUGCAACCUGAGACGCUGCCAACCAGCAGCCG-----CAACAAAAGCAACAACAACUGCGG ((((((((((((((((....))))))..))...((((((....)))))).......))))))))........(((-----((.................))))) ( -25.93) >DroEre_CAF1 78156 104 + 1 UGGCGGUGGCAAAUUGAAAACAAUUUUAGCUAUUUGCAUAUAAAUGCAACCUGAGACGCUGCCAACCAGCAGCAGCCGACCAACAAAAGCUGCCACAACUGCGG .((..(((((((((((....)))))).((((..((((((....))))))........(((((......)))))..............)))))))))..)).... ( -29.80) >DroAna_CAF1 73645 80 + 1 UGGCGGUGGCAAAUUGAAAACAAUUUUAGAUAUUUGCAUAUAAAUGAAACCUGAGAAGCUGCCAACCGGCAGCUG-----CAAUA------------------- ..(((((.(((((((((((....)))))...))))))...........))).....(((((((....))))))))-----)....------------------- ( -20.80) >consensus UGGCGGUGGCAAAUUGAAAACAAUUUUAGCUAUUUGCAUAUAAAUGCAACCUGAGACGCUGCCAACCAGCAGCCG_____CAACAAAAGCAGCAACAACUGCGG .....(((((((((((....))))))..)))))((((((....))))))........(((((......)))))............................... (-14.92 = -15.20 + 0.28)


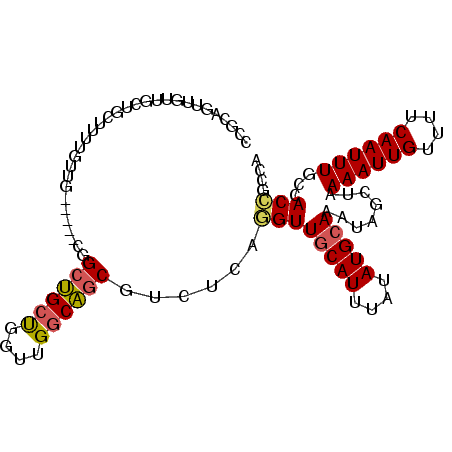
| Location | 7,282,440 – 7,282,539 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.55 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7282440 99 - 20766785 CAGCAGUUGUUGCUGCUUUUGUUG-----CGGCUGCUGGUUGGCAGCGUCUCAAGUUGCAUUUAUAUGCAAAUAGCUAAAAUUGUUUUCAAUUUGCCACCGCCA .(((((((((.((.......)).)-----))))))))((((((((((........((((((....))))))...))).((((((....))))))))))..))). ( -33.10) >DroPse_CAF1 66332 99 - 1 AUCCCUUAGAGCCUCCUCCACCCC-----GUGCCGCCUGCCGCCGCCGUCUCAAGUUGCAUUUAUAUGCAAAUAACUAAAAUUGUUUUCAAUUUGCCACUGUCC ......(((..............(-----(.((.((.....)).)))).......((((((....))))))....)))((((((....)))))).......... ( -13.00) >DroSec_CAF1 78340 99 - 1 CCGCAGUUGUUGCUGCUUUUGUUG-----CGGCUGCUGGUUGGCAGCGUCUCAGGUUGCAUUUAUAUGCAAAUAGCUAAAAUUGUUUUCAAUUUGCCACCGCCA ..((((((((.((.......)).)-----))))))).((((((((((........((((((....))))))...))).((((((....))))))))))..))). ( -32.00) >DroSim_CAF1 81815 99 - 1 CCGCAGUUGUUGUUGCUUUUGUUG-----CGGCUGCUGGUUGGCAGCGUCUCAGGUUGCAUUUAUAUGCAAAUAGCUAAAAUUGUUUUCAAUUUGCCACCGCCA ..((((((((....((....)).)-----))))))).((((((((((........((((((....))))))...))).((((((....))))))))))..))). ( -30.70) >DroEre_CAF1 78156 104 - 1 CCGCAGUUGUGGCAGCUUUUGUUGGUCGGCUGCUGCUGGUUGGCAGCGUCUCAGGUUGCAUUUAUAUGCAAAUAGCUAAAAUUGUUUUCAAUUUGCCACCGCCA ..(((((((.(.((((....)))).)))))))).((.(((.((((((........((((((....))))))...))).((((((....)))))))))))))).. ( -33.70) >DroAna_CAF1 73645 80 - 1 -------------------UAUUG-----CAGCUGCCGGUUGGCAGCUUCUCAGGUUUCAUUUAUAUGCAAAUAUCUAAAAUUGUUUUCAAUUUGCCACCGCCA -------------------....(-----((((((((....))))))).....(((.......((((....))))...((((((....)))))))))...)).. ( -17.70) >consensus CCGCAGUUGUUGCUGCUUUUGUUG_____CGGCUGCUGGUUGGCAGCGUCUCAGGUUGCAUUUAUAUGCAAAUAGCUAAAAUUGUUUUCAAUUUGCCACCGCCA ...............................((((((....))))))......((((((((....)))))........((((((....))))))...))).... (-15.60 = -15.55 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:38 2006