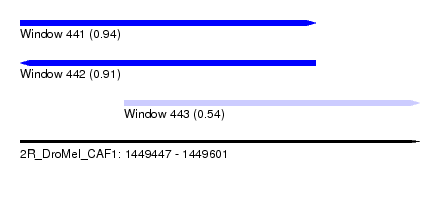
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,449,447 – 1,449,601 |
| Length | 154 |
| Max. P | 0.941492 |

| Location | 1,449,447 – 1,449,561 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.05 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.74 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1449447 114 + 20766785 UGUCUCUUUCAUGGCUGUUAAUUUCCAGCUAUUGUUCGGCCAGCCUCCCGUUUCUGGGC------CCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACG .....((((((((((((.((((........))))..))))))((((.........))))------......))))))(((..((((((.....................))))))..))) ( -29.80) >DroSec_CAF1 67609 114 + 1 UGUCUCUUUCAUGGCUGUUAAUUUCCAGCUAUUGUUCGGCCAGCCUCCCGUGCCUGGGC------CCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACG .....((((((((((((........))))))......((((.(((....).))...)))------).....))))))(((..((((((.....................))))))..))) ( -32.00) >DroSim_CAF1 66485 114 + 1 UGUCUCUUUCAUGGCUGUUAAUUUCCAGCUAUUGUUCGGCCAGCCUCCCGUGCCUGGGC------CCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACG .....((((((((((((........))))))......((((.(((....).))...)))------).....))))))(((..((((((.....................))))))..))) ( -32.00) >DroEre_CAF1 76995 120 + 1 UGUCUCUUUCAUGGCUGUUAAUUUCCAGCUAUUGUUCGGCCAGCCUCCCAUGCCUGGGCCCCGGCCCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACG .....((((((((((((........))))))......((((.(...((((....))))..).)))).....))))))(((..((((((.....................))))))..))) ( -35.60) >DroYak_CAF1 72569 113 + 1 UGUC-CUUUCAUGGCUGUUAAUUUCCAGCUAUUGUUCGGUCAGCCUCCCAUGCCUGGGC------CCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACG ....-((((((.(((((.......(((((....))).)).))))).((((....)))).------......))))))(((..((((((.....................))))))..))) ( -30.70) >consensus UGUCUCUUUCAUGGCUGUUAAUUUCCAGCUAUUGUUCGGCCAGCCUCCCGUGCCUGGGC______CCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACG .....((((((((((((.((((........))))..))))))............((((.......))))..))))))(((..((((((.....................))))))..))) (-29.90 = -29.74 + -0.16)

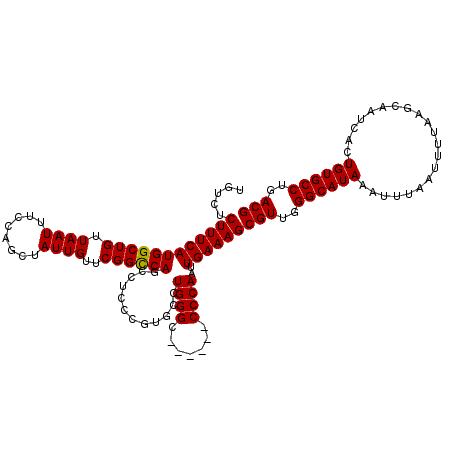
| Location | 1,449,447 – 1,449,561 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.05 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -28.30 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1449447 114 - 20766785 CGUCAGGCACAGUGAUUGCUUAAAAUUAAAUUUAUGCCCAACGCUUUCAAUUGGG------GCCCAGAAACGGGAGGCUGGCCGAACAAUAGCUGGAAAUUAACAGCCAUGAAAGAGACA .(((.((((...(((((......)))))......)))).....((((((((((.(------(((.((...(....).))))))...)))).((((........))))..)))))).))). ( -31.90) >DroSec_CAF1 67609 114 - 1 CGUCAGGCACAGUGAUUGCUUAAAAUUAAAUUUAUGCCCAACGCUUUCAAUUGGG------GCCCAGGCACGGGAGGCUGGCCGAACAAUAGCUGGAAAUUAACAGCCAUGAAAGAGACA .(((.((((...(((((......)))))......)))).....((((((((((.(------(((..(((.(....))))))))...)))).((((........))))..)))))).))). ( -33.70) >DroSim_CAF1 66485 114 - 1 CGUCAGGCACAGUGAUUGCUUAAAAUUAAAUUUAUGCCCAACGCUUUCAAUUGGG------GCCCAGGCACGGGAGGCUGGCCGAACAAUAGCUGGAAAUUAACAGCCAUGAAAGAGACA .(((.((((...(((((......)))))......)))).....((((((((((.(------(((..(((.(....))))))))...)))).((((........))))..)))))).))). ( -33.70) >DroEre_CAF1 76995 120 - 1 CGUCAGGCACAGUGAUUGCUUAAAAUUAAAUUUAUGCCCAACGCUUUCAAUUGGGGCCGGGGCCCAGGCAUGGGAGGCUGGCCGAACAAUAGCUGGAAAUUAACAGCCAUGAAAGAGACA .(((.((((...(((((......)))))......)))).....((((((((((.((((((..((((....))))...))))))...)))).((((........))))..)))))).))). ( -40.40) >DroYak_CAF1 72569 113 - 1 CGUCAGGCACAGUGAUUGCUUAAAAUUAAAUUUAUGCCCAACGCUUUCAAUUGGG------GCCCAGGCAUGGGAGGCUGACCGAACAAUAGCUGGAAAUUAACAGCCAUGAAAG-GACA .(((.((((...(((((......)))))......)))).....((((((.((((.------.((((....)))).......))))......((((........))))..))))))-))). ( -29.00) >consensus CGUCAGGCACAGUGAUUGCUUAAAAUUAAAUUUAUGCCCAACGCUUUCAAUUGGG______GCCCAGGCACGGGAGGCUGGCCGAACAAUAGCUGGAAAUUAACAGCCAUGAAAGAGACA .(((.((((...(((((......)))))......)))).....((((((((((.........(((......))).((....))...)))).((((........))))..)))))).))). (-28.30 = -28.30 + 0.00)

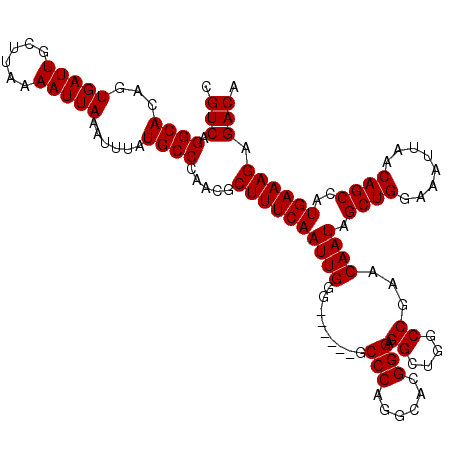
| Location | 1,449,487 – 1,449,601 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1449487 114 + 20766785 CAGCCUCCCGUUUCUGGGC------CCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACGGCUCUUGAUGUCCACGUUGUUGUUUCACGCACACAUCAAU .((((.((((....)))).------.............((..((((((.....................))))))..)))))).(((((((...(((.(......))))...))))))). ( -28.20) >DroSec_CAF1 67649 114 + 1 CAGCCUCCCGUGCCUGGGC------CCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACGGCUCUUGAUGUCCACGUUGUUGUUUCACGCACACAUCAAU .((((.((((....)))).------.............((..((((((.....................))))))..)))))).(((((((...(((.(......))))...))))))). ( -28.20) >DroSim_CAF1 66525 114 + 1 CAGCCUCCCGUGCCUGGGC------CCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACGGCUCUUGAUGUCCACGUUGUUGUUUCACGCACACAUCAAU .((((.((((....)))).------.............((..((((((.....................))))))..)))))).(((((((...(((.(......))))...))))))). ( -28.20) >DroEre_CAF1 77035 120 + 1 CAGCCUCCCAUGCCUGGGCCCCGGCCCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACGGCACUUGAUGUCCACGUUGUUGUUACACGCACACAUCAAU ..........((((.((((....))))...........((..((((((.....................))))))..)))))).(((((((...(((.((....)))))...))))))). ( -34.80) >DroYak_CAF1 72608 114 + 1 CAGCCUCCCAUGCCUGGGC------CCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACGGCACUUGAUGUCCUCGUUGUUGUUACACGCUCACAUCAAA ..(((.((((....)))).------.............((..((((((.....................))))))..)))))..(((((((...(((.((....)))))...))))))). ( -30.30) >consensus CAGCCUCCCGUGCCUGGGC______CCCAAUUGAAAGCGUUGGGCAUAAAUUUAAUUUUAAGCAAUCACUGUGCCUGACGGCUCUUGAUGUCCACGUUGUUGUUUCACGCACACAUCAAU ..(((.........((((.......)))).........((..((((((.....................))))))..)))))..(((((((...(((.(......))))...))))))). (-28.70 = -28.70 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:00 2006