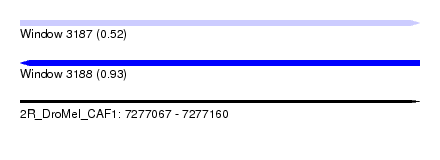
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,277,067 – 7,277,160 |
| Length | 93 |
| Max. P | 0.925528 |

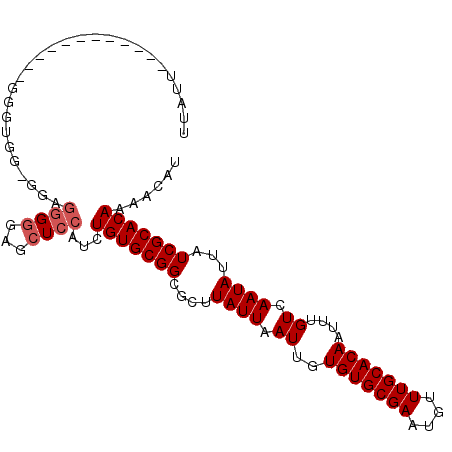
| Location | 7,277,067 – 7,277,160 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -19.27 |
| Energy contribution | -19.77 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
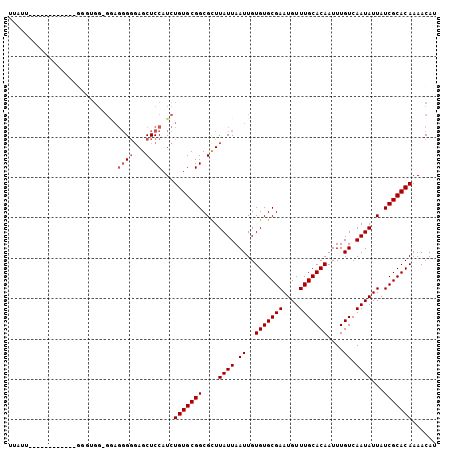
>2R_DroMel_CAF1 7277067 93 + 20766785 AUUUU------------UGGUGG-GGAGGGGGAGCUCCAUCUGUGCGGCGCUUAUUAAUUGUGUGCGAAUGUUUGCACAAUUUGUCAAUAUUAUCGCACAAAACAU .....------------.(((((-((........)))))))(((((((....((((.((..(((((((....)))))))....)).))))...)))))))...... ( -27.10) >DroSec_CAF1 72973 94 + 1 UGAGG------------GGGGGGUGGAGGGGGAGCUCCAUCUGUGCGGCGCUUAUUAAUUGUGUGCGAAUGUUUGCACAAUUUGUCAAUAUUAUCGCACAAAACAU .....------------...((((((((......))))))))((((((....((((.((..(((((((....)))))))....)).))))...))))))....... ( -29.00) >DroSim_CAF1 76452 93 + 1 UGAGG------------GGGAGG-GGAGGGGGAGCUCCAUCUGUGCGGCGCUUAUUAAUUGUGUGCGAAUGUUUGCACAAUUUGUCAAUAUUAUCGCACAAAACAU .....------------.(((..-((((......)))).)))((((((....((((.((..(((((((....)))))))....)).))))...))))))....... ( -25.20) >DroEre_CAF1 73102 105 + 1 UUUUUGUGUUUGGUUUUGUUUGG-GAAGGGGGAGCUCCAUCUGUGCGGCGCUUAUUAAUUGUGUGCGAAUGUUUGCACAAUUUGUCAAUAUUAUCGCACAAAACAU .((((((((..(((..((((.((-.(((...((((.((........)).))))........(((((((....))))))).))).))))))..)))))))))))... ( -25.90) >DroYak_CAF1 74844 104 + 1 UUUUCC-CUUUUUUUGUGUUUGG-GAAGGGGCAGCUCCAUCUGUGCGGCGCUUAUUAAUUGUGUGCGAAUGUUUGCACAAUUUGUCAAUAUUAUCGCACAAAACAU ...(((-((((((........))-)))))))..........(((((((....((((.((..(((((((....)))))))....)).))))...)))))))...... ( -27.10) >DroAna_CAF1 69125 92 + 1 UAAUUCCACUUUUUU--------------GUGAGCUCCAUCUGUGCGGCGUUUAUUAAUUGUGUGCGAAUGUUUGCACAAUUUGUCAAUAUUAUCGCACAAAACAU ......(((......--------------))).........(((((((....((((.((..(((((((....)))))))....)).))))...)))))))...... ( -20.60) >consensus UUAUU____________GGGUGG_GGAGGGGGAGCUCCAUCUGUGCGGCGCUUAUUAAUUGUGUGCGAAUGUUUGCACAAUUUGUCAAUAUUAUCGCACAAAACAU ...........................((((...))))...(((((((....((((.((..(((((((....)))))))....)).))))...)))))))...... (-19.27 = -19.77 + 0.50)

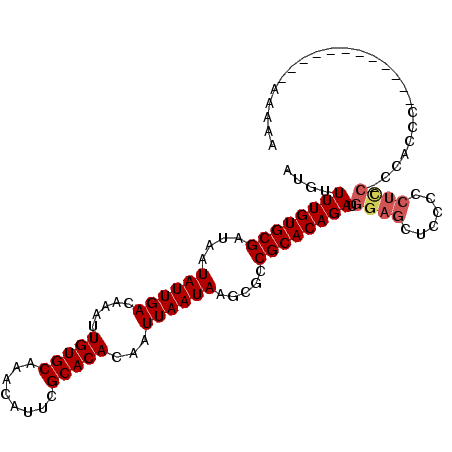
| Location | 7,277,067 – 7,277,160 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -21.17 |
| Consensus MFE | -17.82 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7277067 93 - 20766785 AUGUUUUGUGCGAUAAUAUUGACAAAUUGUGCAAACAUUCGCACACAAUUAAUAAGCGCCGCACAGAUGGAGCUCCCCCUCC-CCACCA------------AAAAU ....((((((((....((((((.....(((((........)))))...)))))).....)))))))).((((......))))-......------------..... ( -21.30) >DroSec_CAF1 72973 94 - 1 AUGUUUUGUGCGAUAAUAUUGACAAAUUGUGCAAACAUUCGCACACAAUUAAUAAGCGCCGCACAGAUGGAGCUCCCCCUCCACCCCCC------------CCUCA ....((((((((....((((((.....(((((........)))))...)))))).....))))))))(((((......)))))......------------..... ( -22.90) >DroSim_CAF1 76452 93 - 1 AUGUUUUGUGCGAUAAUAUUGACAAAUUGUGCAAACAUUCGCACACAAUUAAUAAGCGCCGCACAGAUGGAGCUCCCCCUCC-CCUCCC------------CCUCA ....((((((((....((((((.....(((((........)))))...)))))).....)))))))).((((......))))-......------------..... ( -21.30) >DroEre_CAF1 73102 105 - 1 AUGUUUUGUGCGAUAAUAUUGACAAAUUGUGCAAACAUUCGCACACAAUUAAUAAGCGCCGCACAGAUGGAGCUCCCCCUUC-CCAAACAAAACCAAACACAAAAA .(((((((((((....((((((.....(((((........)))))...)))))).....))))))...((((.......)))-).)))))................ ( -19.10) >DroYak_CAF1 74844 104 - 1 AUGUUUUGUGCGAUAAUAUUGACAAAUUGUGCAAACAUUCGCACACAAUUAAUAAGCGCCGCACAGAUGGAGCUGCCCCUUC-CCAAACACAAAAAAAG-GGAAAA ....((((((((....((((((.....(((((........)))))...)))))).....)))))))).((......)).(((-((.............)-)))).. ( -22.62) >DroAna_CAF1 69125 92 - 1 AUGUUUUGUGCGAUAAUAUUGACAAAUUGUGCAAACAUUCGCACACAAUUAAUAAACGCCGCACAGAUGGAGCUCAC--------------AAAAAAGUGGAAUUA ....((((((((....((((((.....(((((........)))))...)))))).....))))))))..((..((((--------------......))))..)). ( -19.80) >consensus AUGUUUUGUGCGAUAAUAUUGACAAAUUGUGCAAACAUUCGCACACAAUUAAUAAGCGCCGCACAGAUGGAGCUCCCCCUCC_CCACCC____________AAAAA ....((((((((....((((((.....(((((........)))))...)))))).....)))))))).((((......))))........................ (-17.82 = -18.32 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:34 2006