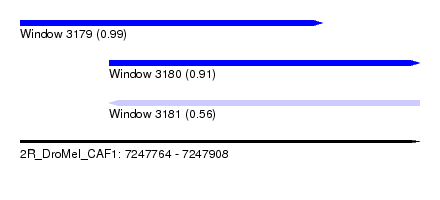
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,247,764 – 7,247,908 |
| Length | 144 |
| Max. P | 0.989954 |

| Location | 7,247,764 – 7,247,873 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.36 |
| Mean single sequence MFE | -18.04 |
| Consensus MFE | -12.90 |
| Energy contribution | -12.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7247764 109 + 20766785 UCUCGAAUGUGAUAUUCUCAGACCCCUCAAACUCCUCCCCGCC--CCCCGUAAAAAAAAAAAAACUUAGGUGUCAAUAUUCCGUUCUAUUGACGCCCUCGAACAAUAACCC ..((((.(.(((.....))).).....................--.......................((((((((((........)))))))))).)))).......... ( -16.20) >DroPse_CAF1 37748 85 + 1 ---------GGGGUUUGUCGCCCCCCUCCUCCUACCCCCUCGU---CCC--------------AGUGAGGUGUCAAUAUCCCGUUCUAUUGACGCCCUCGAACAAUGGCCC ---------(((((.....)))))...................---.((--------------(.(((((((((((((........)))))))).))))).....)))... ( -24.60) >DroSec_CAF1 43286 103 + 1 UCUCGAAUGUGAUAUUCUCAGACCCCUACCAUUUCCCUCUCCC---CCC-----AAAAAAAAGACUGAGGUGUCAAUAUUCCGUUCUAUUGACGCCCUCGAACAAUAACCC ..((((...........((((......................---...-----..........))))((((((((((........)))))))))).)))).......... ( -16.55) >DroSim_CAF1 45880 106 + 1 UCUCGAAUGUGAUAUUCUCAGACCCCUACCAUUUCCCUCUCCCCCCCCC-----AAAAAAAAGACUGAGGUGUCAAUAUUCCGUUCUAUUGACGCCCUCGAACAAUAACCC ..((((...........((((............................-----..........))))((((((((((........)))))))))).)))).......... ( -16.46) >DroYak_CAF1 44961 94 + 1 UCUCGAAUGCGAUAUUCUCAGACCCCUACCUUUUU-----UC----CCC--------AAAACGACUAAGGUGUCAAUAUUCCGUUCUAUUGACGCCCUCGAACAAUAACCC ..((((..((.................(((((...-----((----...--------.....))..)))))(((((((........)))))))))..)))).......... ( -16.50) >DroAna_CAF1 41183 94 + 1 -CCCCAAGUCGAAAUUCUUAACCCCCUCCA---UC-----GC----GCCAAUACCUAAUAUAAUGUGAGGUGUCAAUAUUCCGUUCUAUUGACGCCCUCGAACAAUG---- -.......((((..................---((-----((----(...(((.....)))..)))))((((((((((........)))))))))).))))......---- ( -17.90) >consensus UCUCGAAUGUGAUAUUCUCAGACCCCUACCAUUUCCC_CUCCC___CCC_____AAAAAAAAGACUGAGGUGUCAAUAUUCCGUUCUAUUGACGCCCUCGAACAAUAACCC ....................................................................((((((((((........))))))))))............... (-12.90 = -12.90 + 0.00)

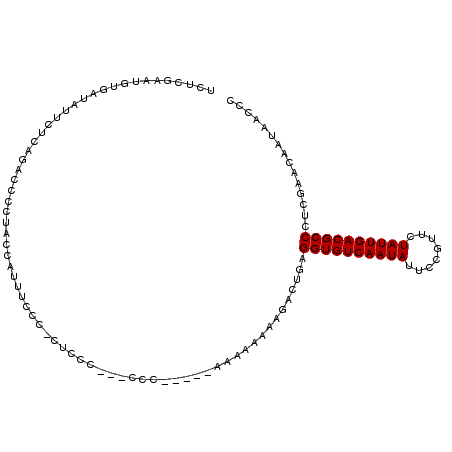
| Location | 7,247,796 – 7,247,908 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -15.13 |
| Energy contribution | -15.30 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7247796 112 + 20766785 UCCUCCCCGCC--CCCCGUAAAAAAAAAAAAACUUAGGUGUCAAUAUUCCGUUCUAUUGACGCCCUCGAACAAUAACCCGAAACA--AACUUCAACAACUUGUCCCCGGCACUCGA ........(((--.......................((((((((((........))))))))))...............(((...--...)))..............)))...... ( -18.50) >DroSec_CAF1 43318 106 + 1 UUCCCUCUCCC---CCC-----AAAAAAAAGACUGAGGUGUCAAUAUUCCGUUCUAUUGACGCCCUCGAACAAUAACCCGAAACA--AACUUCAAAAACUUGUCCCCGGCACUCGA ...........---...-----........((((((((((((((((........)))))))).)))))...........(((...--...)))........)))............ ( -17.00) >DroSim_CAF1 45912 109 + 1 UUCCCUCUCCCCCCCCC-----AAAAAAAAGACUGAGGUGUCAAUAUUCCGUUCUAUUGACGCCCUCGAACAAUAACCCGAAACA--AACUUCAAAAACUUGUCCCCGGCACUCGA .................-----........((((((((((((((((........)))))))).)))))...........(((...--...)))........)))............ ( -17.00) >DroEre_CAF1 42548 94 + 1 CUC-----GC----ACC--------AAAA---CUGAGGUGUCAAUAUGCCGUUCUAUUGACGCCCUCGAACAAUAACCCGAAACA--AACUUCAAAAACUUGUGCCCGGCACUCGA ...-----((----((.--------....---.(((((((((((((........)))))))).)))))...........(((...--...)))........))))........... ( -19.80) >DroYak_CAF1 44993 99 + 1 UUU-----UC----CCC--------AAAACGACUAAGGUGUCAAUAUUCCGUUCUAUUGACGCCCUCGAACAAUAACCCGAAACAAAAACUUCAAAAACUUGUCCCCGGCACUCGA ...-----..----...--------....(((....((((((((((........)))))))))).............(((..((((.............))))...)))...))). ( -17.12) >DroPer_CAF1 37662 94 + 1 UACCCCCUCGU---CCC--------------AGUGAGGUGUCAAUAUCCCGUUCUAUUGACGCCCUCGAACAAUGGCCCGAA-CA--UA-UUCA-AAUCCAGUCUCCGGCACUCGU .......(((.---.((--------------(.(((((((((((((........)))))))).))))).....)))..))).-..--..-....-......((.....))...... ( -21.90) >consensus UUCCC_CUCCC___CCC________AAAAAGACUGAGGUGUCAAUAUUCCGUUCUAUUGACGCCCUCGAACAAUAACCCGAAACA__AACUUCAAAAACUUGUCCCCGGCACUCGA ....................................((((((((((........)))))))))).((((..........(((........)))........((.....))..)))) (-15.13 = -15.30 + 0.17)

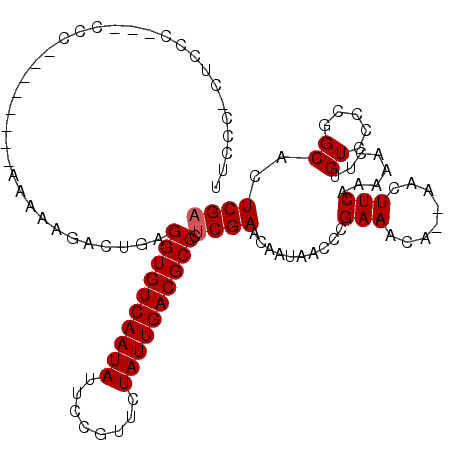

| Location | 7,247,796 – 7,247,908 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -21.74 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7247796 112 - 20766785 UCGAGUGCCGGGGACAAGUUGUUGAAGUU--UGUUUCGGGUUAUUGUUCGAGGGCGUCAAUAGAACGGAAUAUUGACACCUAAGUUUUUUUUUUUUUACGGGG--GGCGGGGAGGA (((((.(((.(..((((..(.....)..)--)))..).))).....)))))(((.(((((((........))))))).)))..........(((((..((...--..))..))))) ( -29.50) >DroSec_CAF1 43318 106 - 1 UCGAGUGCCGGGGACAAGUUUUUGAAGUU--UGUUUCGGGUUAUUGUUCGAGGGCGUCAAUAGAACGGAAUAUUGACACCUCAGUCUUUUUUUU-----GGG---GGGAGAGGGAA .(((..(((.(..((((..(.....)..)--)))..).)))..)))...((((..(((((((........))))))).))))..(((((((((.-----...---.))))))))). ( -31.10) >DroSim_CAF1 45912 109 - 1 UCGAGUGCCGGGGACAAGUUUUUGAAGUU--UGUUUCGGGUUAUUGUUCGAGGGCGUCAAUAGAACGGAAUAUUGACACCUCAGUCUUUUUUUU-----GGGGGGGGGAGAGGGAA .(((..(((.(..((((..(.....)..)--)))..).)))..)))...((((..(((((((........))))))).))))..((((((((((-----......)))))))))). ( -31.50) >DroEre_CAF1 42548 94 - 1 UCGAGUGCCGGGCACAAGUUUUUGAAGUU--UGUUUCGGGUUAUUGUUCGAGGGCGUCAAUAGAACGGCAUAUUGACACCUCAG---UUUU--------GGU----GC-----GAG ....(..(((((.((((...(((((((..--..)))))))...)))).)((((..(((((((........))))))).))))..---..))--------)).----.)-----... ( -27.90) >DroYak_CAF1 44993 99 - 1 UCGAGUGCCGGGGACAAGUUUUUGAAGUUUUUGUUUCGGGUUAUUGUUCGAGGGCGUCAAUAGAACGGAAUAUUGACACCUUAGUCGUUUU--------GGG----GA-----AAA (((((.(((.(..(((((...........)))))..).))).....)))))....(((((((........))))))).((((((.....))--------)))----).-----... ( -26.50) >DroPer_CAF1 37662 94 - 1 ACGAGUGCCGGAGACUGGAUU-UGAA-UA--UG-UUCGGGCCAUUGUUCGAGGGCGUCAAUAGAACGGGAUAUUGACACCUCACU--------------GGG---ACGAGGGGGUA .(..((.((((....(((.((-((((-..--..-)))))))))......((((..(((((((........))))))).)))).))--------------)).---))..)...... ( -29.20) >consensus UCGAGUGCCGGGGACAAGUUUUUGAAGUU__UGUUUCGGGUUAUUGUUCGAGGGCGUCAAUAGAACGGAAUAUUGACACCUCAGUCUUUUU________GGG___GGGAG_GGGAA .......((..((((((...(((((((......)))))))...))))))((((..(((((((........))))))).)))).................))............... (-21.74 = -22.53 + 0.79)

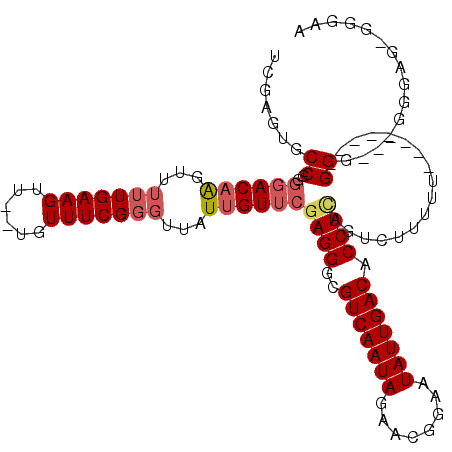
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:28 2006