
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,204,528 – 7,204,663 |
| Length | 135 |
| Max. P | 0.955891 |

| Location | 7,204,528 – 7,204,629 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.08 |
| Mean single sequence MFE | -20.78 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.94 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
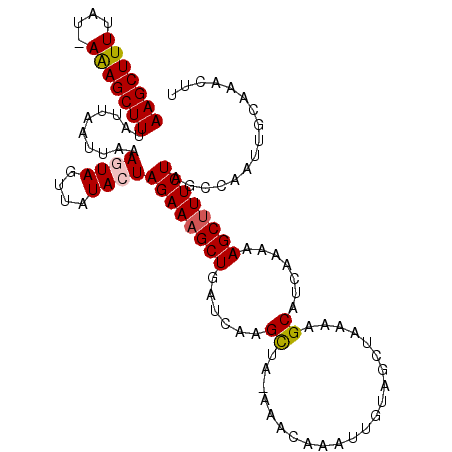
>2R_DroMel_CAF1 7204528 101 + 20766785 UUUCACAUUUUCACACUUUUUAAAAAAGUUCGCAAUUGGCGAGAAAGCUUUUUGAUGCUUUUAGCUACAAUUUGUUU-UGGCUUGAUCAGCUUUCUAGUAUA ..............(((((.....)))))((((.....))))(((((((..(..(.(((...(((........))).-.))))..)..)))))))....... ( -20.60) >DroSec_CAF1 4185 93 + 1 UUUCACAUUUUCACACUUUUCUAA-AAGUAUGCA-------AGAAAGCUUUUUGAUGCUUUUAUAUACAAUUUGUUU-UAGCUUGAUCAGCGUUCUAGUAUA ..............(((((....)-))))((((.-------((((.(((..(..(.(((...(((.......)))..-.))))..)..))).)))).)))). ( -15.60) >DroSim_CAF1 7045 100 + 1 UUUCACAUUUUCACACUUUUUGAA-AAGUUCGCAAUUGGCGAGAAAGCUUUUUGAUGCUUUUAGCUACAAUUUGUUU-UAGCUUGAUCAGCUUUCUAGUAUA .......((((((.......))))-))..((((.....))))(((((((..(..(.(((...(((........))).-.))))..)..)))))))....... ( -22.00) >DroEre_CAF1 6147 100 + 1 CAUCGAUAUCCCGCACGUU-UUAAAUGUUUUGCAAAUGGCUAGAAAGCUUUUUGAUGCUUUUAGCUAAAAUUUGCUU-CAACUCGAUCAGCUUUCUAGUAUU ............((((((.-....))))...)).....(((((((((((..((((.(.((..(((........))).-.)))))))..)))))))))))... ( -22.80) >DroYak_CAF1 7590 96 + 1 ------AAUCCCGCACUUUUUUAAAAAAUUGGCAAGUGGCUAGAAAGCUUUUUGAUGCUUUUUGCUAACAAUUUUUUAAGGCUUGAUCAGCUUUCUAGUAUU ------.......(((((..((((....)))).)))))(((((((((((..(..(.(((((................))))))..)..)))))))))))... ( -22.89) >consensus UUUCACAUUUUCACACUUUUUUAAAAAGUUCGCAAUUGGCGAGAAAGCUUUUUGAUGCUUUUAGCUACAAUUUGUUU_UAGCUUGAUCAGCUUUCUAGUAUA ......................................((.((((((((..((((.(((...(((........)))...)))))))..)))))))).))... (-12.86 = -13.94 + 1.08)

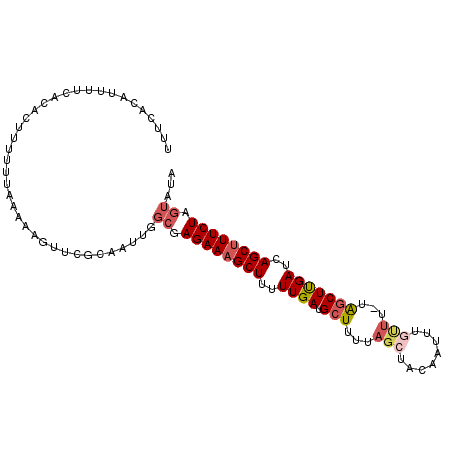
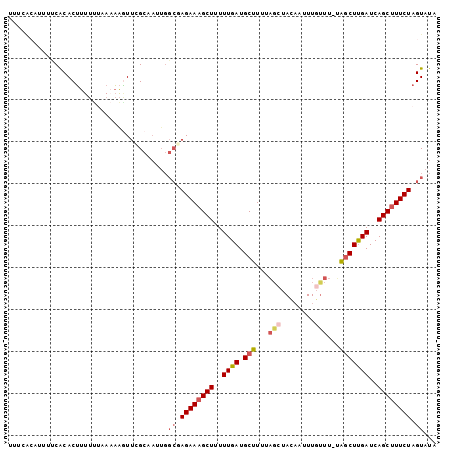
| Location | 7,204,553 – 7,204,663 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -15.62 |
| Energy contribution | -17.10 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7204553 110 + 20766785 AAGUUCGCAAUUGGCGAGAAAGCUUUUUGAUGCUUUUAGCUACAAUUUGUUU-UGGCUUGAUCAGCUUUCUAGUAUAACUAAUUUAAUUAAUAAAGCUCUCAUAAAAGCUU .(((((((.....)))((((((((..(..(.(((...(((........))).-.))))..)..)))))))).....)))).............(((((........))))) ( -23.30) >DroSec_CAF1 4209 91 + 1 AAGUAUGCA-------AGAAAGCUUUUUGAUGCUUUUAUAUACAAUUUGUUU-UAGCUUGAUCAGCGUUCUAGUAUAACUACUUUAACUAAAAAAG------------CUU .(((((((.-------((((.(((..(..(.(((...(((.......)))..-.))))..)..))).)))).)))).)))................------------... ( -16.20) >DroSim_CAF1 7069 110 + 1 AAGUUCGCAAUUGGCGAGAAAGCUUUUUGAUGCUUUUAGCUACAAUUUGUUU-UAGCUUGAUCAGCUUUCUAGUAUAACUAUUUUAACUAAAAAAGCUUUAAUAAAAGCUU .(((((((.....)))((((((((..(..(.(((...(((........))).-.))))..)..)))))))).....)))).............(((((((....))))))) ( -25.40) >DroEre_CAF1 6171 110 + 1 UGUUUUGCAAAUGGCUAGAAAGCUUUUUGAUGCUUUUAGCUAAAAUUUGCUU-CAACUCGAUCAGCUUUCUAGUAUUACUACUUUAAUUAAUAAAGCUUUGAUAAAAGCUU .............(((((((((((..((((.(.((..(((........))).-.)))))))..)))))))))))...................(((((((....))))))) ( -27.30) >DroYak_CAF1 7609 111 + 1 AAAUUGGCAAGUGGCUAGAAAGCUUUUUGAUGCUUUUUGCUAACAAUUUUUUAAGGCUUGAUCAGCUUUCUAGUAUUACUACUUUAAUUAAUAAAGCUUUUAUAAAAGCUU .((((((..(((((((((((((((..(..(.(((((................))))))..)..)))))))))))..))))...))))))....(((((((....))))))) ( -27.59) >consensus AAGUUCGCAAUUGGCGAGAAAGCUUUUUGAUGCUUUUAGCUACAAUUUGUUU_UAGCUUGAUCAGCUUUCUAGUAUAACUACUUUAAUUAAUAAAGCUUU_AUAAAAGCUU .............((.((((((((..((((.(((...(((........)))...)))))))..)))))))).))...................(((((((....))))))) (-15.62 = -17.10 + 1.48)

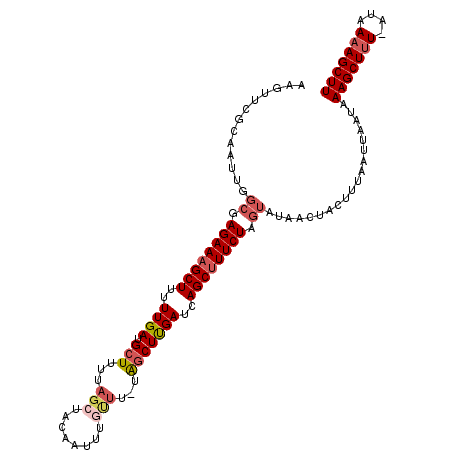
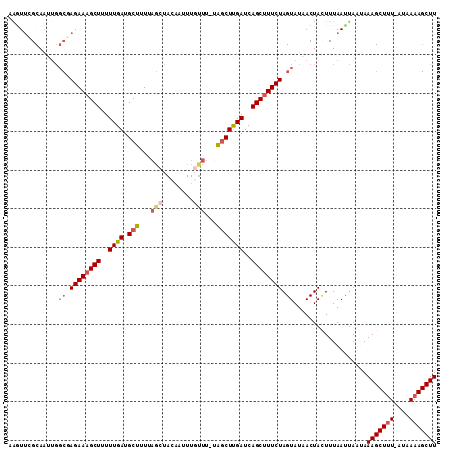
| Location | 7,204,553 – 7,204,663 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -21.19 |
| Consensus MFE | -10.67 |
| Energy contribution | -11.19 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7204553 110 - 20766785 AAGCUUUUAUGAGAGCUUUAUUAAUUAAAUUAGUUAUACUAGAAAGCUGAUCAAGCCA-AAACAAAUUGUAGCUAAAAGCAUCAAAAAGCUUUCUCGCCAAUUGCGAACUU ((((((((.(((..(((((...........((((((((.......(((.....)))..-........))))))))))))).)))))))))))..((((.....)))).... ( -21.93) >DroSec_CAF1 4209 91 - 1 AAG------------CUUUUUUAGUUAAAGUAGUUAUACUAGAACGCUGAUCAAGCUA-AAACAAAUUGUAUAUAAAAGCAUCAAAAAGCUUUCU-------UGCAUACUU .((------------(((..(((((...((((....)))).....)))))..))))).-........((((....(((((........)))))..-------))))..... ( -16.00) >DroSim_CAF1 7069 110 - 1 AAGCUUUUAUUAAAGCUUUUUUAGUUAAAAUAGUUAUACUAGAAAGCUGAUCAAGCUA-AAACAAAUUGUAGCUAAAAGCAUCAAAAAGCUUUCUCGCCAAUUGCGAACUU ((((((((.....(((((..((((((....(((.....)))...))))))..))))).-............((.....))....))))))))..((((.....)))).... ( -23.40) >DroEre_CAF1 6171 110 - 1 AAGCUUUUAUCAAAGCUUUAUUAAUUAAAGUAGUAAUACUAGAAAGCUGAUCGAGUUG-AAGCAAAUUUUAGCUAAAAGCAUCAAAAAGCUUUCUAGCCAUUUGCAAAACA (((((((....)))))))..............((((..((((((((((....(((((.-.(((........)))...))).))....))))))))))....))))...... ( -25.70) >DroYak_CAF1 7609 111 - 1 AAGCUUUUAUAAAAGCUUUAUUAAUUAAAGUAGUAAUACUAGAAAGCUGAUCAAGCCUUAAAAAAUUGUUAGCAAAAAGCAUCAAAAAGCUUUCUAGCCACUUGCCAAUUU (((((((....)))))))..............((((..((((((((((...(((...........)))...((.....)).......))))))))))....))))...... ( -18.90) >consensus AAGCUUUUAU_AAAGCUUUAUUAAUUAAAGUAGUUAUACUAGAAAGCUGAUCAAGCUA_AAACAAAUUGUAGCUAAAAGCAUCAAAAAGCUUUCUAGCCAAUUGCAAACUU (((((((....)))))))..........((((....))))((((((((......((......................)).......))))))))................ (-10.67 = -11.19 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:18 2006