
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,186,380 – 7,186,495 |
| Length | 115 |
| Max. P | 0.780238 |

| Location | 7,186,380 – 7,186,495 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -16.44 |
| Energy contribution | -16.96 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7186380 115 + 20766785 AAAGCAGGCAUUAAUCAAAUAUUUCUUAGAACUGAUAAGUUCAGCGAAAAAGGUGUCACUCAAUACCAAGAAAGAUGACAAAAUGCGAUUAGCUCUUUGUU-----GUUUUUCAAUCGAA ............................(((((....)))))..(((....(((((......)))))..((((((..(((((..((.....))..))))).-----.))))))..))).. ( -27.60) >DroSec_CAF1 19439 114 + 1 AAAGCGGGCAUUAAUCAAAUAUUUCUUGGAACUGAUAAGUUCAGUGAA-AAGGUGUCACUCAAUACCAAGAAAGAUGACAAAAUGCGAUUAGCUCUUUGUU-----GUUUUCCAAUCUAA .....(((.............((((((((((((....)))))(((((.-......)))))......)))))))((..(((((..((.....))..))))).-----.)).)))....... ( -26.30) >DroSim_CAF1 18075 114 + 1 AAAGCAGGCAUUAAUCAAAUAUUUCUUGGAACUGAUAAGUUCAGUGGA-AAGGUGUCACUCAAUACCAAGAAAGAUGACAAAAUGCGAUUAGCUCUUUGUU-----GUUUUUCAAUCUAA .....................(((((..(((((....)))))...)))-))(((((......)))))..((((((..(((((..((.....))..))))).-----.))))))....... ( -29.30) >DroEre_CAF1 17194 112 + 1 AGGGCUGUCAUUAAUCA--CCUUUCUUGGAACAGAUAAGUUCGGUGAA-GGGGUGUCACUCGAUACCAAGAAAGAUUACAAAAGGCGAUUGGCUUCUUGGU-----GUUUUUCAAAGUAA .(((.((.(((......--((((((.(.((((......)))).).)))-)))))).)))))(((((((((((.(((..(.....)..)))...))))))))-----)))........... ( -29.70) >DroYak_CAF1 18393 119 + 1 AAAUUUAUCGUUAAUCAAACAGUUCUUGGAACUGAUAAGUUCAGUGAA-AGGCUGUCACUCAAUACCAAGAAAGAUCACGAAACGCGAUUGGCUUUUUGUUUAUUCGAUUAACAAAAUAG .........(((((((......(((((((((((....)))))(((((.-......)))))......)))))).((..((((((.((.....)).))))))....)))))))))....... ( -27.90) >consensus AAAGCAGGCAUUAAUCAAAUAUUUCUUGGAACUGAUAAGUUCAGUGAA_AAGGUGUCACUCAAUACCAAGAAAGAUGACAAAAUGCGAUUAGCUCUUUGUU_____GUUUUUCAAUCUAA (((((................((((((((((((....)))))(((((........)))))......)))))))...((((((..((.....))..)))))).....)))))......... (-16.44 = -16.96 + 0.52)

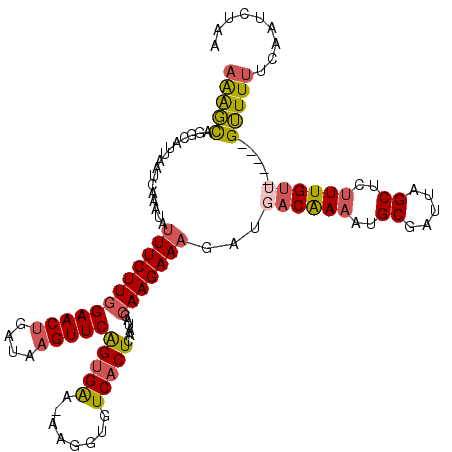
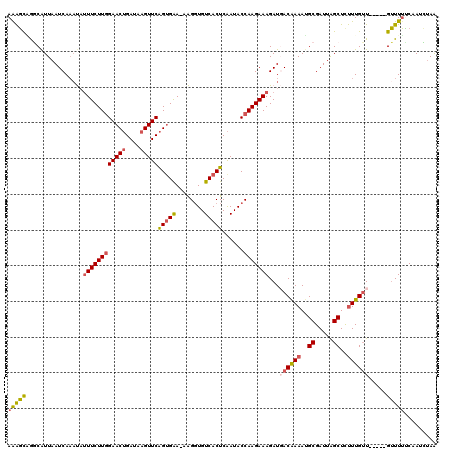
| Location | 7,186,380 – 7,186,495 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.06 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7186380 115 - 20766785 UUCGAUUGAAAAAC-----AACAAAGAGCUAAUCGCAUUUUGUCAUCUUUCUUGGUAUUGAGUGACACCUUUUUCGCUGAACUUAUCAGUUCUAAGAAAUAUUUGAUUAAUGCCUGCUUU ..............-----.....(((((.....(((((..((((..((((((((.(((((((((........))))........))))).))))))))....)))).)))))..))))) ( -22.60) >DroSec_CAF1 19439 114 - 1 UUAGAUUGGAAAAC-----AACAAAGAGCUAAUCGCAUUUUGUCAUCUUUCUUGGUAUUGAGUGACACCUU-UUCACUGAACUUAUCAGUUCCAAGAAAUAUUUGAUUAAUGCCCGCUUU ...((((((.....-----.........))))))(((((..((((..((((((((.(((((((((......-.))))........))))).))))))))....)))).)))))....... ( -25.74) >DroSim_CAF1 18075 114 - 1 UUAGAUUGAAAAAC-----AACAAAGAGCUAAUCGCAUUUUGUCAUCUUUCUUGGUAUUGAGUGACACCUU-UCCACUGAACUUAUCAGUUCCAAGAAAUAUUUGAUUAAUGCCUGCUUU ..............-----.....(((((.....(((((..((((..((((((((.((((((((.......-..)))........))))).))))))))....)))).)))))..))))) ( -23.40) >DroEre_CAF1 17194 112 - 1 UUACUUUGAAAAAC-----ACCAAGAAGCCAAUCGCCUUUUGUAAUCUUUCUUGGUAUCGAGUGACACCCC-UUCACCGAACUUAUCUGUUCCAAGAAAGG--UGAUUAAUGACAGCCCU .............(-----(((((((.((.....)).)))))....(((((((((..(((.((((......-.)))))))((......)).))))))))))--))............... ( -22.30) >DroYak_CAF1 18393 119 - 1 CUAUUUUGUUAAUCGAAUAAACAAAAAGCCAAUCGCGUUUCGUGAUCUUUCUUGGUAUUGAGUGACAGCCU-UUCACUGAACUUAUCAGUUCCAAGAACUGUUUGAUUAACGAUAAAUUU .....((((((((((((((............((((((...))))))..(((((((.(((((....(((...-....)))......))))).))))))).))))))))))))))....... ( -31.20) >consensus UUAGAUUGAAAAAC_____AACAAAGAGCUAAUCGCAUUUUGUCAUCUUUCUUGGUAUUGAGUGACACCUU_UUCACUGAACUUAUCAGUUCCAAGAAAUAUUUGAUUAAUGCCAGCUUU ....................((((((.((.....)).))))))....((((((((.(((((((((........))))........))))).))))))))..................... (-15.78 = -16.06 + 0.28)

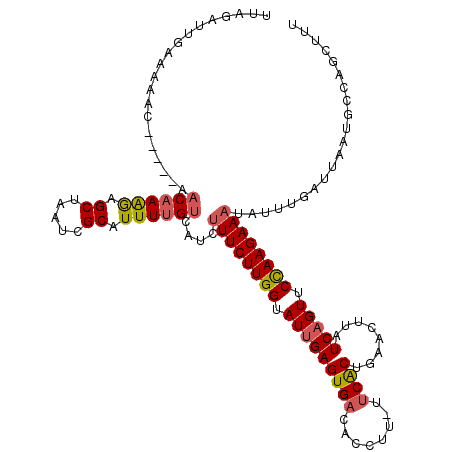
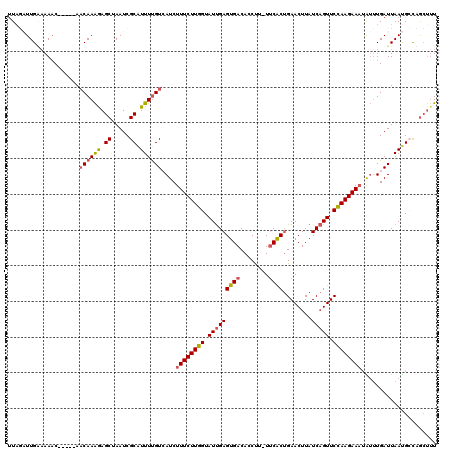
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:07 2006