
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,173,499 – 7,173,597 |
| Length | 98 |
| Max. P | 0.861963 |

| Location | 7,173,499 – 7,173,597 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.13 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
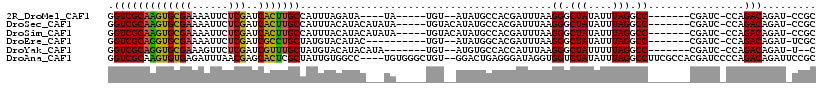
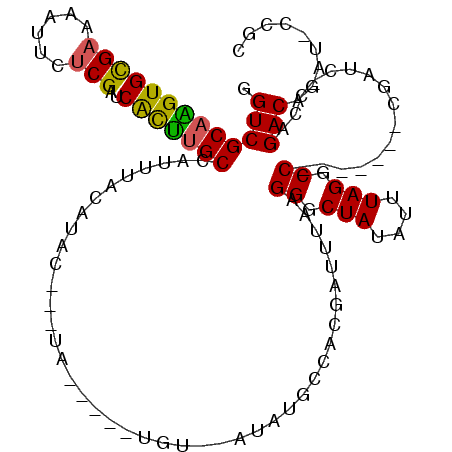
>2R_DroMel_CAF1 7173499 98 - 20766785 GGUCGCAAGUGCGAAAAUUCUCGAUCACUUGCCAUUUAGAUA----UA-----UGU--AUAUGCCACGAUUUAAGGGCUAUAUUUAGGCC-------CGAUC-CCAGACAGAU-CCGC ((((((((((((((......)))..)))))))..........----((-----.((--(((.(((.(.......))))))))).))))))-------.((((-.......)))-)... ( -24.60) >DroSec_CAF1 6962 104 - 1 GGUCGCAAGUGCGAAAAUUCUCGAUCACUUGCCAUUUACAUACAUAUA-----UGUACAUAUGCCACGAUUUAAGGGCUAUAUUUAGGCC-------CGAUC-CCAGACAGAU-CCGC (((.((((((((((......)))..)))))))....((((((....))-----)))).....))).((......(((((.......))))-------)((((-.......)))-))). ( -27.20) >DroSim_CAF1 5623 104 - 1 GGUCGCAAGUGCGAAAAUUCUCGAUCACUUGCCAUUUACAUACAUAUA-----UGUACAUAUGCCACGAUUUAAGGGCUAUAUUUAGGCC-------CGAUC-CCAGACAGAU-CCGC (((.((((((((((......)))..)))))))....((((((....))-----)))).....))).((......(((((.......))))-------)((((-.......)))-))). ( -27.20) >DroEre_CAF1 4591 97 - 1 GGUCGCAGGUGCGAAAAUUCUCGAUCGCCUGCUAUGUACAUAC----------UGU--AUAUGGCACGAUUUAAGGGCUAUAUUUAGGCC-------CGAUC-CCAGACAGAU-UCGC ((((((....))))........((((.(.((((((((((....----------.))--)))))))).)......(((((.......))))-------)))))-))........-.... ( -31.20) >DroYak_CAF1 5580 98 - 1 GGUCGCAGGUGCGAAAGUUCUCGAUCGUUUGCUAUGUACAUACAUA-------UGU--AUGUGCCACCAUUUAAGGGCUAUUUUUAGGCC-------CGAUC-CCAGACAGAU-U--C (((((.(((.((....))))))))))(((((....((((((((...-------.))--))))))..........(((((.......))))-------)....-.)))))....-.--. ( -32.70) >DroAna_CAF1 4514 112 - 1 GGUCGCAAGUGUGAGAUUUAACGAGCACUCGCUAUUGUGGCC----UGUGGGCUGU--GGACUGAGGGAUAGGUGGUCUAUAUUUAGGCCUUCGCCACGAUCCCCAGACAGAUUCCGC ((((((((..(((((............)))))..))))))))----.((((..(.(--(..(((.(((((.(((((((((....))))))...)))...)))))))).)).)..)))) ( -44.80) >consensus GGUCGCAAGUGCGAAAAUUCUCGAUCACUUGCCAUUUACAUAC___UA_____UGU__AUAUGCCACGAUUUAAGGGCUAUAUUUAGGCC_______CGAUC_CCAGACAGAU_CCGC .(((((((((((((......)))..)))))))..........................................((.(((....))).))................)))......... (-17.66 = -17.13 + -0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:03 2006