
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,140,087 – 7,140,211 |
| Length | 124 |
| Max. P | 0.988764 |

| Location | 7,140,087 – 7,140,189 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -41.09 |
| Consensus MFE | -26.25 |
| Energy contribution | -26.42 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
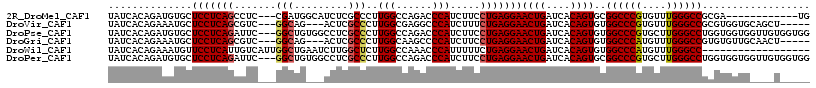
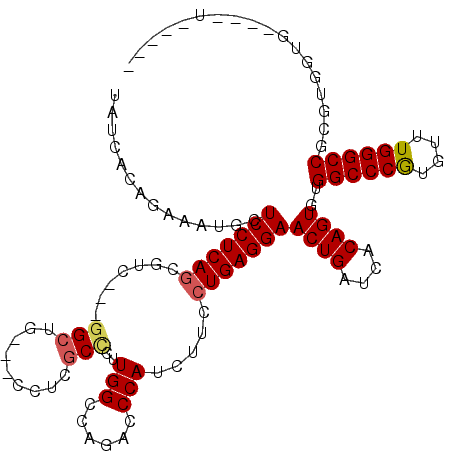
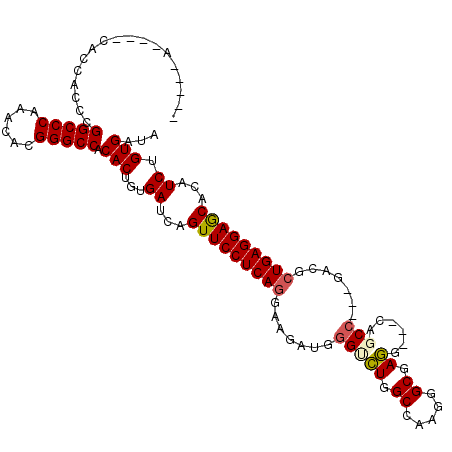
>2R_DroMel_CAF1 7140087 102 + 20766785 UAUCACAGAUGUGCUCCUCAGCCUC---CGAUGGCAUCUCGCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGCGGCCCGUGUUUGGGCCGCGA------------UG ((((...(((..(.(((((((....---.(((((..(((.(((...))).))).)))))...))))))).)..))).....((((((((....))))))))))------------)) ( -43.30) >DroVir_CAF1 4803 106 + 1 UAUCACAGAAAUGCUCCUCAGCGUC---GGCAG---ACUCGCCCUUGGCGAGGCCCAUCUUUCUGAGGAACUGAUCACAGUGUGGCCCGUGUUUGGGCCGCGUGGUGCAGCU----- ((((((........(((((((.((.---(((..---.((((((...))))))))).))....)))))))((((....))))((((((((....)))))))))))))).....----- ( -45.90) >DroPse_CAF1 3344 114 + 1 UAUCACAGAUGUGCUCCUCAGAUUC---GGCUGUGGCCUCGCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGUGGCCCGUGCUUGGGCCUGGUGGUGGUUGUGGUGG ((((((........(((((((....---((.(.(((((........))))).).))......)))))))...((((((.....((((((....)))))).....)))))))))))). ( -46.40) >DroGri_CAF1 4827 106 + 1 UAUCACAGAAAUGCUCCUCAGCGUC---GGCAG---ACUCGCCCUUGGCAAGCCCCAUCUUCCUGAGGAACUGAUCACAGUGUGGCCCAUGUUUGGGCCGUGUGUUGCAACU----- ...........((((((((((.(((---....)---))..(((...))).............)))))))......((((...(((((((....)))))))))))..)))...----- ( -34.50) >DroWil_CAF1 3909 99 + 1 UAUCACAGAAAUGUUCCUCAUUGUCAUUGGCUGAAUCUUGGCUCUUGGCCAAACCCAUUUUUCUGAGGAACUGAUCACAGUGUGGCCCAUGUUUGGGCC------------------ ...(((.((...((((((((.......(((.......(((((.....)))))..)))......))))))))...))...))).((((((....))))))------------------ ( -30.72) >DroPer_CAF1 3341 114 + 1 UAUCACAGAUGUGCUCCUCAGAUUC---GGCUGUGGCCUCGCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGCGGCCCGUGCUUGGGCCUGGUGGUGGUUGUGGUGG ((((((........(((((((....---((.(.(((((........))))).).))......)))))))...((((((.....((((((....)))))).....)))))))))))). ( -45.70) >consensus UAUCACAGAAAUGCUCCUCAGCGUC___GGCUG___CCUCGCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGUGGCCCGUGUUUGGGCCGCGUGGUG____U_____ ..............(((((((.......(((.........)))..(((......))).....)))))))((((....))))..((((((....)))))).................. (-26.25 = -26.42 + 0.17)
| Location | 7,140,087 – 7,140,189 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -45.46 |
| Consensus MFE | -32.06 |
| Energy contribution | -32.22 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
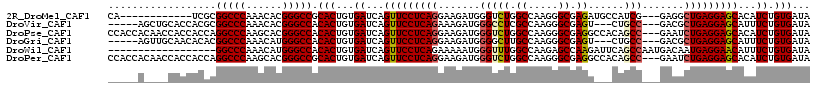
>2R_DroMel_CAF1 7140087 102 - 20766785 CA------------UCGCGGCCCAAACACGGGCCGCACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGCGAGAUGCCAUCG---GAGGCUGAGGAGCACAUCUGUGAUA .(------------(((((((((......)))))))((...(((..(((((((((...(((((((((.(((...))).)))).))))).---....)))))))))..))).))))). ( -50.40) >DroVir_CAF1 4803 106 - 1 -----AGCUGCACCACGCGGCCCAAACACGGGCCACACUGUGAUCAGUUCCUCAGAAAGAUGGGCCUCGCCAAGGGCGAGU---CUGCC---GACGCUGAGGAGCAUUUCUGUGAUA -----.((........))(((((......))))).(((...((...(((((((((...(.(.(((((((((...)))))).---..)))---.)).)))))))))...)).)))... ( -45.30) >DroPse_CAF1 3344 114 - 1 CCACCACAACCACCACCAGGCCCAAGCACGGGCCACACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGCGAGGCCACAGCC---GAAUCUGAGGAGCACAUCUGUGAUA ....((((..(((.....(((((......))))).....)))....((((((((((..(.(((((((.(((...))).))))).))..)---...)))))))))).....))))... ( -46.80) >DroGri_CAF1 4827 106 - 1 -----AGUUGCAACACACGGCCCAAACAUGGGCCACACUGUGAUCAGUUCCUCAGGAAGAUGGGGCUUGCCAAGGGCGAGU---CUGCC---GACGCUGAGGAGCAUUUCUGUGAUA -----.((..((.((((.((((((....))))))....))))....(((((((((...(.(((((((((((...)))))))---)..))---).).))))))))).....))..)). ( -46.20) >DroWil_CAF1 3909 99 - 1 ------------------GGCCCAAACAUGGGCCACACUGUGAUCAGUUCCUCAGAAAAAUGGGUUUGGCCAAGAGCCAAGAUUCAGCCAAUGACAAUGAGGAACAUUUCUGUGAUA ------------------((((((....)))))).(((...((...((((((((........((((((((.....)))))......)))........))))))))...)).)))... ( -36.19) >DroPer_CAF1 3341 114 - 1 CCACCACAACCACCACCAGGCCCAAGCACGGGCCGCACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGCGAGGCCACAGCC---GAAUCUGAGGAGCACAUCUGUGAUA ....((((..(((.....(((((......))))).....)))....((((((((((..(.(((((((.(((...))).))))).))..)---...)))))))))).....))))... ( -47.90) >consensus _____A____CACCACCCGGCCCAAACACGGGCCACACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGCGAGG___CAGCC___GACGCUGAGGAGCACAUCUGUGAUA ..................(((((......))))).(((...((...(((((((((.......(((((.((.....)).))......))).......)))))))))...)).)))... (-32.06 = -32.22 + 0.17)
| Location | 7,140,121 – 7,140,211 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.71 |
| Mean single sequence MFE | -37.68 |
| Consensus MFE | -24.04 |
| Energy contribution | -25.27 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
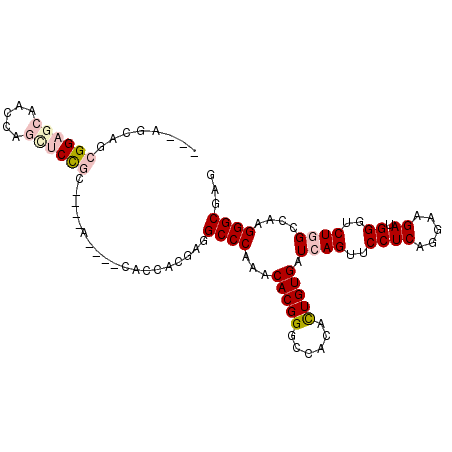
>2R_DroMel_CAF1 7140121 90 - 20766785 ---AGCAGCGGAGCAAGCAGCUCCGCA------------UCGCGGCCCAAACACGGGCCGCACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGCGAG ---.((.(((((((.....))))))).------------..(((((((......))))))).((.((..(((..((((.....)).))..)))..)).))))... ( -44.50) >DroVir_CAF1 4834 96 - 1 UGCGGAAACGG---AACCAGCUCC------AGCUGCACCACGCGGCCCAAACACGGGCCACACUGUGAUCAGUUCCUCAGAAAGAUGGGCCUCGCCAAGGGCGAG .(((....)((---...((((...------.))))..))..))(((((...(((((......)))))(((..(((....))).))))))))(((((...))))). ( -35.90) >DroPse_CAF1 3378 102 - 1 ---AGCAGCGGAGCAAGUAGUUCUGCCACCACAACCACCACCAGGCCCAAGCACGGGCCACACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGCGAG ---.((.(((((((.....)))))))........((....(((((((((..(((((......))))).....((((...))))..)))))))))....))))... ( -38.20) >DroMoj_CAF1 4340 96 - 1 ACUAGUUCAGG---UUCCAGCUCC------AGUUGCACCACACGGCCCAAACACGGGCCACACUGUGAUCAGUUCCUCAGAAAGAUGGGCCUCGCCAAGGGCGAC .........((---(..((((...------.)))).)))....(((((...(((((......)))))(((..(((....))).))))))))(((((...))))). ( -30.60) >DroAna_CAF1 3220 90 - 1 ---AGCAACGGAGCCACUGGCUCCGCA------------UCGAGGCCCAAACACGGGCCGCAUUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGCGAG ---.....(((((((...)))))))..------------((..(((((......)))))((....((..(((..((((.....)).))..)))..))...)))). ( -36.30) >DroPer_CAF1 3375 102 - 1 ---AGCAGCGGAGCAAGCAGCUCUGCCACCACAACCACCACCAGGCCCAAGCACGGGCCGCACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGCGAG ---.((.(((((((.....)))))))........((....(((((((((..(((((......))))).....((((...))))..)))))))))....))))... ( -40.60) >consensus ___AGCAGCGGAGCAACCAGCUCCGC____A____CACCACGAGGCCCAAACACGGGCCACACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGCGAG ........((((((.....))))))...................((((...(((((......))))).((((..((((.....)).))..))))....))))... (-24.04 = -25.27 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:50 2006