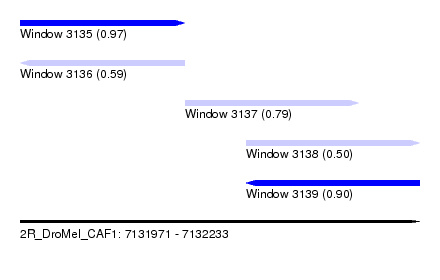
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,131,971 – 7,132,233 |
| Length | 262 |
| Max. P | 0.973938 |

| Location | 7,131,971 – 7,132,079 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.16 |
| Mean single sequence MFE | -45.92 |
| Consensus MFE | -36.92 |
| Energy contribution | -37.48 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7131971 108 + 20766785 CUCUUCGAUUUUCGCUGCCUGCAGUGCUGCCGCU------------GCACGAGUGUGAGUGCAUGUUAGUCGCUGGCAGAGCUAUGUUAACUAACAGUGGCGAGAGCGUGGCGGCGAAAC .........(((((((((((((....((((((((------------((((........)))))(((((((.((((((...)))).))..)))))))))))).)).))).)))))))))). ( -44.70) >DroSec_CAF1 159448 108 + 1 CCCUUCGAUUUUCGCUGCCUGCCGCGCUGCCGCU------------GCACGAGUGUGAGUGCAUGUUAGUCGCUGGCAGAGCUAUGUUAACUAACAGAGGCGAGAGCGUGGCGGCGAAAC .........((((((((((...(((.(((((..(------------((((........)))))(((((((.((((((...)))).))..)))))))..))).)).))).)))))))))). ( -44.30) >DroSim_CAF1 157750 108 + 1 CUCUUCGAUUUUCGCUGCCUGCCGUGCUGCCGCU------------GCACGAGUGUGAGUGCAUGUUAGUCGCUGGCAGAGCUAUGUUAACUAACAGUGGCGAGAGCGUGGCGGCGAAAC .........((((((((((...(((.((((((((------------((((........)))))(((((((.((((((...)))).))..)))))))))))).)).))).)))))))))). ( -45.60) >DroEre_CAF1 165055 120 + 1 CUCUUCGAUUUUCGCUGCCUGCAGUUCUGCCGCUGCCGCUGCCACUGCACGAGUGUGAGUGCAUGUUAGGCGCUGGCAGAGCCAUGUUAACUAACAGAGAUGAGAGCGGAGCGGCGAAAC .........(((((((((((((.(((((((((.((((...((...(((((........))))).))..)))).)))))))))(((.((........)).)))...)))).))))))))). ( -51.50) >DroYak_CAF1 169102 108 + 1 CUCUUCGAUUUUCGCUGCCUGCAGUUCUGCCGCU------------GCACGAGUGUGAGUGCAUGUUAGUCGCUGGCAGAGCUAUGUUAACUAACAGCGAUGAGAGUGAGGCGGCGAAAC .........(((((((((((((((((((((((((------------((((........)))))........)).))))))))).((((....)))))).((....)).))))))))))). ( -43.50) >consensus CUCUUCGAUUUUCGCUGCCUGCAGUGCUGCCGCU____________GCACGAGUGUGAGUGCAUGUUAGUCGCUGGCAGAGCUAUGUUAACUAACAGAGGCGAGAGCGUGGCGGCGAAAC .........(((((((((((((....(((((((.............((((........)))).(((((((.((((((...)))).))..)))))))))))).)).))).)))))))))). (-36.92 = -37.48 + 0.56)

| Location | 7,131,971 – 7,132,079 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.16 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -28.48 |
| Energy contribution | -28.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7131971 108 - 20766785 GUUUCGCCGCCACGCUCUCGCCACUGUUAGUUAACAUAGCUCUGCCAGCGACUAACAUGCACUCACACUCGUGC------------AGCGGCAGCACUGCAGGCAGCGAAAAUCGAAGAG .((((((.(((.(((.........((((((((......(((.....)))))))))))(((((........))))------------))))(((....))).))).))))))......... ( -34.90) >DroSec_CAF1 159448 108 - 1 GUUUCGCCGCCACGCUCUCGCCUCUGUUAGUUAACAUAGCUCUGCCAGCGACUAACAUGCACUCACACUCGUGC------------AGCGGCAGCGCGGCAGGCAGCGAAAAUCGAAGGG .((((((.(((.(((.((.(((..((((((((......(((.....)))))))))))(((((........))))------------)..))))).)))...))).))))))......... ( -38.60) >DroSim_CAF1 157750 108 - 1 GUUUCGCCGCCACGCUCUCGCCACUGUUAGUUAACAUAGCUCUGCCAGCGACUAACAUGCACUCACACUCGUGC------------AGCGGCAGCACGGCAGGCAGCGAAAAUCGAAGAG .((((((.(((.((..((.(((..((((((((......(((.....)))))))))))(((((........))))------------)..)))))..))...))).))))))......... ( -34.70) >DroEre_CAF1 165055 120 - 1 GUUUCGCCGCUCCGCUCUCAUCUCUGUUAGUUAACAUGGCUCUGCCAGCGCCUAACAUGCACUCACACUCGUGCAGUGGCAGCGGCAGCGGCAGAACUGCAGGCAGCGAAAAUCGAAGAG .(((((((((((((((.((((...((((((......((((...))))....))))))(((((........))))))))).))))).))))))....(((....))).))))......... ( -44.00) >DroYak_CAF1 169102 108 - 1 GUUUCGCCGCCUCACUCUCAUCGCUGUUAGUUAACAUAGCUCUGCCAGCGACUAACAUGCACUCACACUCGUGC------------AGCGGCAGAACUGCAGGCAGCGAAAAUCGAAGAG .((((((.((((((........(((((........)))))((((((.(((.....).(((((........))))------------)))))))))..)).)))).))))))......... ( -35.70) >consensus GUUUCGCCGCCACGCUCUCGCCACUGUUAGUUAACAUAGCUCUGCCAGCGACUAACAUGCACUCACACUCGUGC____________AGCGGCAGCACUGCAGGCAGCGAAAAUCGAAGAG .((((((.(((.((((........((((((((......(((.....))))))))))).((((........))))............))))((......)).))).))))))......... (-28.48 = -28.72 + 0.24)

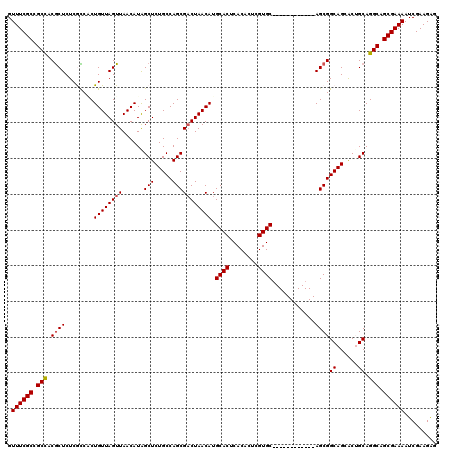
| Location | 7,132,079 – 7,132,193 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.02 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -17.79 |
| Energy contribution | -18.03 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7132079 114 + 20766785 AAGUGGACAUGAAACCAAAUCUUGCUGUGUAGUUGUGGGCACAAACAAACAAACACGCACACUUGCACACGGUUCUAUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGA ...((((..(((...........(((((((.((.(((.((................)).)))..))))))))).....(((((((....))))))))))))))........... ( -21.59) >DroSec_CAF1 159556 114 + 1 AAGUGGAAAUGAAACCAAAUCUUACCGUGUAGUUGUGGGCACAAACAAACAAACACGCACACAUGCACACGGCUCUGUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGA ...((((.................((((((.((((((.((................)).)))).))))))))...((.(((((((....))))))).))))))........... ( -24.99) >DroSim_CAF1 157858 114 + 1 AAGUGGAAAUGAAACCAAAUCUUACCGUGUAGUUGUGGGCACAAACAAACAAACACGCACACAUGCACACGGCUCUGUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGA ...((((.................((((((.((((((.((................)).)))).))))))))...((.(((((((....))))))).))))))........... ( -24.99) >DroEre_CAF1 165175 108 + 1 AACUGGAA-UGAAACCAAAUCUUGC-AUGUAGUUGUGGGCACAAAC----AAACACGCACACAUGCACACGCUUCUGUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGA ...((((.-.............(((-((((.((((((.........----...)))).)))))))))........((.(((((((....))))))).))))))........... ( -22.90) >DroYak_CAF1 169210 110 + 1 AUGUGGAAUUGAAACAAAAUCUUGCCGUGUAUUUGUGUGCACAAAC----AAACACACACACUUGCGCACGCUUCUGUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGA ...((((................((.(((((..((((((.......----.....))))))..)))))..))...((.(((((((....))))))).))))))........... ( -23.90) >consensus AAGUGGAAAUGAAACCAAAUCUUGCCGUGUAGUUGUGGGCACAAACAAACAAACACGCACACAUGCACACGGUUCUGUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGA ...((((...................(((((..((((.(................).))))..))))).......((.(((((((....))))))).))))))........... (-17.79 = -18.03 + 0.24)

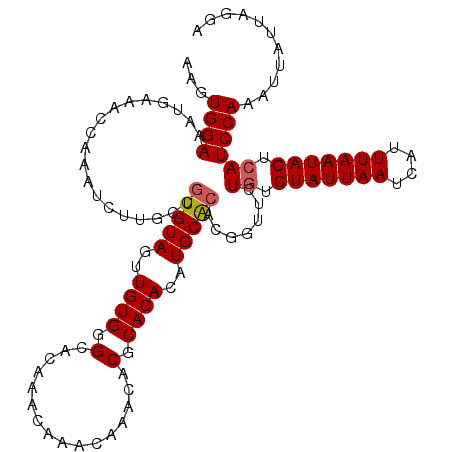
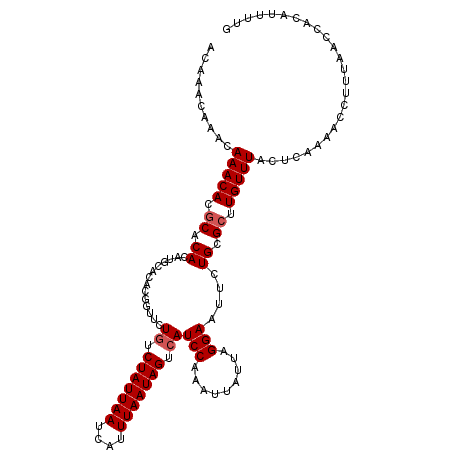
| Location | 7,132,119 – 7,132,233 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -16.84 |
| Consensus MFE | -12.18 |
| Energy contribution | -12.58 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7132119 114 + 20766785 ACAAACAAACAAACACGCACACUUGCACACGGUUCUAUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGAAUUCUGCGCUUGUUUAGACAAAACCUUUAACCACAUUUUG ......((((((...((((...................(((((((....)))))))...(((.........)))....)))).))))))......................... ( -12.70) >DroSec_CAF1 159596 114 + 1 ACAAACAAACAAACACGCACACAUGCACACGGCUCUGUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGAAUUUUGAGCUUGUUUACUCAAAACCUUUAACCACAUUUUG ......((((((....(((....))).....((((((.(((((((....))))))).))(((.........))).....))))))))))......................... ( -18.50) >DroSim_CAF1 157898 114 + 1 ACAAACAAACAAACACGCACACAUGCACACGGCUCUGUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGAAUUUUGCGCUUGUUUACUCAAAACCUUUAACCACAUUUUG ......((((((...((((.....((.....))..((.(((((((....))))))).))(((.........)))....)))).))))))......................... ( -15.10) >DroEre_CAF1 165213 110 + 1 ACAAAC----AAACACGCACACAUGCACACGCUUCUGUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGAAUUCUGCGCUUGUUUGCGCAAAACCUUUAACCACAUUUUG ......----.....((((.(((.((.((......((.(((((((....))))))).))(((.........)))....)).)).))).))))...................... ( -19.20) >DroYak_CAF1 169250 109 + 1 ACAAAC----AAACACACACACUUGCGCACGCUUCUGUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGAAUUCUGCGCUUGUUUGCUCAAAACCUUUAACCACAUU-CG .(((((----((............(((((......((.(((((((....))))))).))(((.........)))....)))))))))))).....................-.. ( -18.70) >consensus ACAAACAAACAAACACGCACACAUGCACACGGUUCUGUCUAUUAAUCAUUUAAUAGUCAUCCAAAUUAUUAGGAAUUCUGCGCUUGUUUACUCAAAACCUUUAACCACAUUUUG ..........(((((.((.((..............((.(((((((....))))))).))(((.........)))....)).)).)))))......................... (-12.18 = -12.58 + 0.40)

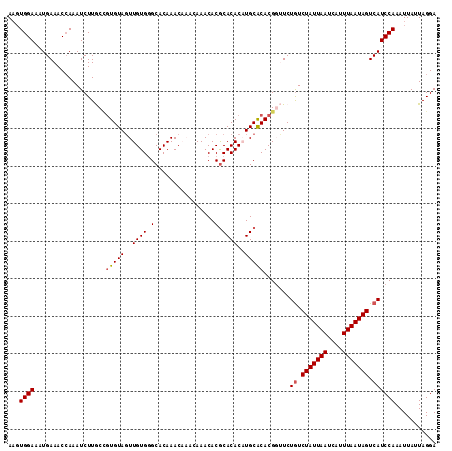
| Location | 7,132,119 – 7,132,233 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7132119 114 - 20766785 CAAAAUGUGGUUAAAGGUUUUGUCUAAACAAGCGCAGAAUUCCUAAUAAUUUGGAUGACUAUUAAAUGAUUAAUAGAUAGAACCGUGUGCAAGUGUGCGUGUUUGUUUGUUUGU ((((((.(......).))))))..(((((((((((.....(((.........)))...(((((((....)))))))..........(..(....)..))))))))))))..... ( -23.70) >DroSec_CAF1 159596 114 - 1 CAAAAUGUGGUUAAAGGUUUUGAGUAAACAAGCUCAAAAUUCCUAAUAAUUUGGAUGACUAUUAAAUGAUUAAUAGACAGAGCCGUGUGCAUGUGUGCGUGUUUGUUUGUUUGU ((....(((((((...(((((((((......)))))))))(((.........))))))))))....))...((((((((((..((..((....))..))..))))))))))... ( -27.00) >DroSim_CAF1 157898 114 - 1 CAAAAUGUGGUUAAAGGUUUUGAGUAAACAAGCGCAAAAUUCCUAAUAAUUUGGAUGACUAUUAAAUGAUUAAUAGACAGAGCCGUGUGCAUGUGUGCGUGUUUGUUUGUUUGU ((((((.(......).))))))..(((((((((((.................((.((.(((((((....))))))).))...))..(..(....)..))))))))))))..... ( -26.60) >DroEre_CAF1 165213 110 - 1 CAAAAUGUGGUUAAAGGUUUUGCGCAAACAAGCGCAGAAUUCCUAAUAAUUUGGAUGACUAUUAAAUGAUUAAUAGACAGAAGCGUGUGCAUGUGUGCGUGUUU----GUUUGU ((....(((((((...(((((((((......)))))))))(((.........))))))))))....))....(((((((((.(((..((....))..))).)))----)))))) ( -31.00) >DroYak_CAF1 169250 109 - 1 CG-AAUGUGGUUAAAGGUUUUGAGCAAACAAGCGCAGAAUUCCUAAUAAUUUGGAUGACUAUUAAAUGAUUAAUAGACAGAAGCGUGCGCAAGUGUGUGUGUUU----GUUUGU ..-....................((((((((((((.....(((.........)))((.(((((((....))))))).))...(((..(....)..)))))))))----)))))) ( -28.40) >consensus CAAAAUGUGGUUAAAGGUUUUGAGUAAACAAGCGCAGAAUUCCUAAUAAUUUGGAUGACUAUUAAAUGAUUAAUAGACAGAACCGUGUGCAUGUGUGCGUGUUUGUUUGUUUGU ((....(((((((...(((((((((......)))))))))(((.........))))))))))....))....(((((((((...(..((((....))))..)...))))))))) (-22.04 = -22.52 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:47 2006