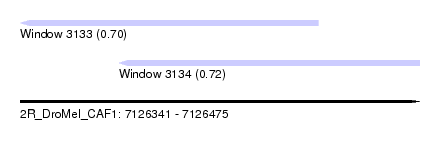
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,126,341 – 7,126,475 |
| Length | 134 |
| Max. P | 0.720468 |

| Location | 7,126,341 – 7,126,441 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -14.95 |
| Energy contribution | -15.41 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
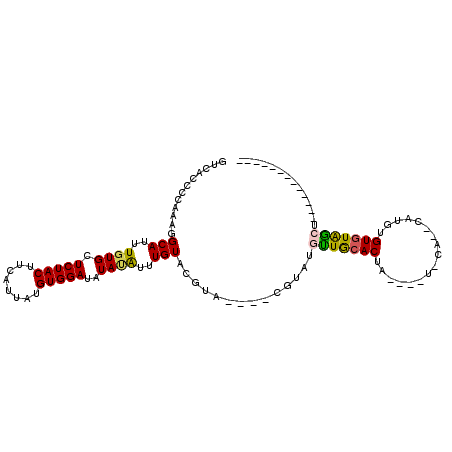
>2R_DroMel_CAF1 7126341 100 - 20766785 UGUGGAUAUAUAUUUGUACGUACGUAUGUUGCACUAGUACA--CAUGUGUAUAGCUGUAUAGCUGUAUAGCUGUGUGUGCACGGCAGGCGACUCUGUGUGUG .(((.(((((((.(((((((((((((((((((....))).)--)))))))))((((....)))))))))).))))))).))).(((.(((....))).))). ( -31.50) >DroSec_CAF1 153964 89 - 1 UGUGGAUAUAUAUUUGUACGUACGUAUGUUGCACUACUACACACAUGUGUGUAG-----UAGCUGU------GUGUGUACACGACAGGCGUCUCUGU--GUG .(((.((((((....((((....))))...(((((((((((((....)))))))-----))).)))------)))))).))).(((((....)))))--... ( -28.60) >DroEre_CAF1 159576 77 - 1 UGUGGAUAUACAUUUGUACGUACGUAUGCUGCACCA---CA--CAUGUGUGUAGCU------------------GUGUGCGCGGCAGGCGUCUUUGU--GUG ......((((((...(.((((.....((((((..((---((--((.((.....)))------------------))))).)))))).)))))..)))--))) ( -25.30) >consensus UGUGGAUAUAUAUUUGUACGUACGUAUGUUGCACUA_UACA__CAUGUGUGUAGCU___UAGCUGU______GUGUGUGCACGGCAGGCGUCUCUGU__GUG .(((.((((((....((((....))))(((((((..............))))))).................)))))).))).(((((....)))))..... (-14.95 = -15.41 + 0.45)



| Location | 7,126,374 – 7,126,475 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.47 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.45 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7126374 101 - 20766785 GUCACCCCAAAGGCAUUUGUGCUCUACUUCAUUAUGUGGAUAUAUAUUUGUACGUA----CGUAUGUUGCACUA---GUACA--CAUGUGUAUAGCUGUAUAGCUGUAUA ............((((.((((((((((........)))))........(((((((.----...))).))))...---)))))--.))))((((((((....)))))))). ( -23.10) >DroSec_CAF1 153992 95 - 1 GUCACCCCAAAGGCAUUUGUGCUCUACUUCAUUAUGUGGAUAUAUAUUUGUACGUA----CGUAUGUUGCACUA---CUACACACAUGUGUGUAG-----UAGCUGU--- ...........(((((.((((((((((........)))))((((....)))).)))----)).)))))((((((---(((((((....)))))))-----))).)))--- ( -29.00) >DroEre_CAF1 159602 85 - 1 GUCACCCCAAAGGCAUUUGUGCUCUACUUCAUUAUGUGGAUAUACAUUUGUACGUA----CGUAUGCUGCACCA------CA--CAUGUGUGUAGCU------------- ...........(((((.((((((((((........)))))..(((....))).)))----)).)))))((..((------((--....))))..)).------------- ( -22.00) >DroYak_CAF1 163535 89 - 1 GUCACCCCAAAGGCAUUUGUGCUCUACUUCAUUAUGUGGAUAUAUAUUUGUACGUAUCUAUGUAUGUUGCACUA------CA--CAUGUGUGUAGCU------------- ...........(((....((((...((.....((((((((((..((....))..)))))))))).)).))))((------((--(....))))))))------------- ( -20.70) >DroAna_CAF1 155692 83 - 1 GCCACCCCGAAGGCAUUUCUGCUCUACUUCAUUAUGUGGAUAUAUGUAUGUACA------------AUACACAACUUCUCCA--CGUGUGUGUGUGU------------- (((((...(((((((....)))....))))..((((((((....(((((.....------------))))).......))))--)))).))).))..------------- ( -17.80) >consensus GUCACCCCAAAGGCAUUUGUGCUCUACUUCAUUAUGUGGAUAUAUAUUUGUACGUA____CGUAUGUUGCACUA____U_CA__CAUGUGUGUAGCU_____________ ............(((..((((.(((((........)))))..))))..)))..............(((((((.................))))))).............. (-12.69 = -12.45 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:42 2006