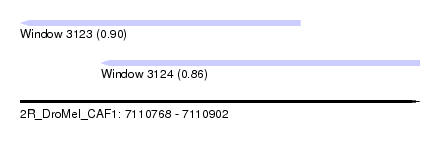
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,110,768 – 7,110,902 |
| Length | 134 |
| Max. P | 0.899602 |

| Location | 7,110,768 – 7,110,862 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.92 |
| Mean single sequence MFE | -16.04 |
| Consensus MFE | -7.06 |
| Energy contribution | -7.66 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7110768 94 - 20766785 ACAAUAAUUACUAGAUUUCUUAUCGGAAAUCUUCAGUAAAACAUCCUGACUCAUCUUCGUAAUUACG-----------------UUU--A-------CCUAACAACCUUUGAAAUACAUU .((((((((((.((((((((....))))))))((((.........)))).........))))))).(-----------------((.--.-------...))).....)))......... ( -13.10) >DroSec_CAF1 138721 103 - 1 AUAAUAAUUACUAGAUUUCGUAUCGGAAAUCUUCAGUAAAACUUGCUGAUUCAUCUUCGUAAUUACA-----------------GCUGUAUCUCAAUACUAACAACCUUUGAAAUACAUU ....(((((((.(((((((......)))))))((((((.....)))))).........)))))))..-----------------..(((((.((((............)))).))))).. ( -20.80) >DroSim_CAF1 138742 94 - 1 AUAAUAAUUACUAGAUUUCUUAUCGGAAAUCUUCAGUAAAACUUGCUGAUUCAUCUUCGUAAUUACA-----------------GCUGUAUCUCAAUACUAACAACCUUUG--------- ....(((((((.((((((((....))))))))((((((.....)))))).........)))))))..-----------------...((((....))))............--------- ( -16.60) >DroEre_CAF1 143898 120 - 1 ACAAUAAUUACUAGUUUUCUUAUCGGAAAUCUUCAGCAAAAAAUACCGACUCAUCUUGGCAAUUACAUUUGGAAGCACAUCGUGGCUGUAUCUCAACACUAACAACAUUUGAAAUACAUG .......((((..((((((.....))))))((((...........((((......))))(((......)))))))......)))).(((((.((((............)))).))))).. ( -11.60) >DroYak_CAF1 148061 108 - 1 ACAAUAAUUACUAGAUUUCUUAUCGGAAAUCUUCAGUAUAACAUCCCAACUCAUCUUCGCA------------AGCACAUCGUAGCUGUAUCUCAACACUAACAGCAGUUGAAACAUAUU ........((((((((((((....))))))))..))))........(((((......(...------------.).........(((((............))))))))))......... ( -18.10) >consensus ACAAUAAUUACUAGAUUUCUUAUCGGAAAUCUUCAGUAAAACAUCCUGACUCAUCUUCGUAAUUACA_________________GCUGUAUCUCAACACUAACAACCUUUGAAAUACAUU .......((((((((((((......)))))))..)))))................................................................................. ( -7.06 = -7.66 + 0.60)

| Location | 7,110,795 – 7,110,902 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -7.56 |
| Energy contribution | -9.72 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7110795 107 - 20766785 UCAAGUGCAUUACAACAACGGAAAACUCUUUUCCAUGGAUACAAUAAUUACUAGAUUUCUUAUCGGAAAUCUUCAGUAAAACAUCCUGACUCAUCUUCGUAAUUACG------------- ....(((.(((((......(((((.....)))))..((((.......(((((((((((((....))))))))..)))))...))))............)))))))).------------- ( -20.90) >DroSec_CAF1 138757 107 - 1 UCAAGUGCAUUACAACAACGGAAAACUCUUUUCCGUGGAUAUAAUAAUUACUAGAUUUCGUAUCGGAAAUCUUCAGUAAAACUUGCUGAUUCAUCUUCGUAAUUACA------------- ....(((.(((((....(((((((.....)))))))((((............(((((((......)))))))((((((.....))))))...))))..)))))))).------------- ( -25.00) >DroSim_CAF1 138769 107 - 1 UCAAGUGCAUUACAACAGCGGAAAACUCUUUUCCAUGGAUAUAAUAAUUACUAGAUUUCUUAUCGGAAAUCUUCAGUAAAACUUGCUGAUUCAUCUUCGUAAUUACA------------- ....(((.(((((......(((((.....)))))(((((.............((((((((....))))))))((((((.....)))))))))))....)))))))).------------- ( -22.70) >DroEre_CAF1 143938 119 - 1 UCAAGUGCAUUUCAACUUCGAAAAACUC-UAGCAACAGAUACAAUAAUUACUAGUUUUCUUAUCGGAAAUCUUCAGCAAAAAAUACCGACUCAUCUUGGCAAUUACAUUUGGAAGCACAU ....((((.((((((.(((((......(-(((..................))))........)))))..................((((......)))).........)))))))))).. ( -15.51) >DroYak_CAF1 148101 107 - 1 UCAAGUGCAUUACAACUUAGAAAAUCCC-AACCAACAGAUACAAUAAUUACUAGAUUUCUUAUCGGAAAUCUUCAGUAUAACAUCCCAACUCAUCUUCGCA------------AGCACAU ....((((...............(((..-........)))........((((((((((((....))))))))..)))).......................------------.)))).. ( -13.90) >consensus UCAAGUGCAUUACAACAACGGAAAACUCUUUUCCAUGGAUACAAUAAUUACUAGAUUUCUUAUCGGAAAUCUUCAGUAAAACAUCCUGACUCAUCUUCGUAAUUACA_____________ .....(((...........(((((.....))))).............((((((((((((......)))))))..)))))...................)))................... ( -7.56 = -9.72 + 2.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:33 2006