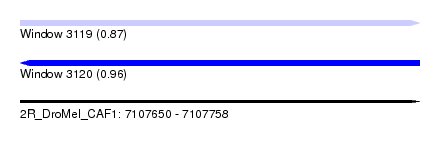
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,107,650 – 7,107,758 |
| Length | 108 |
| Max. P | 0.963541 |

| Location | 7,107,650 – 7,107,758 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -55.45 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.50 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
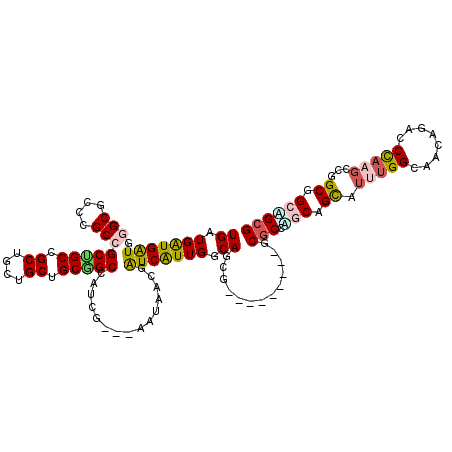
>2R_DroMel_CAF1 7107650 108 + 20766785 UGGUGGUGAUGGGCUCCCGCCGCAGCCGCCGCUGCUGCUGCCAUGG---AAUAACGGUCAUUGGCAGAG---------GCGCCUCCAGCUUGUGGCAAUAAACCCAAUCCGGCAGCAGCG .(((((((.(((((....))).))..)))))))(((((((((..((---(.....(((.((((.((.((---------((.......)))).)).))))..)))...)))))))))))). ( -51.60) >DroVir_CAF1 174016 114 + 1 UGGUGAUGGUGGGCGCCAGCAGCUGCCGCAGCUGCUGCGGCCAUUGAUGAGUAACGAUCAUUGGCAGCC------GCUGCGCCGGCAGCAUUUGGUAAUAGGCCCAAGCCGGCGGCCGCG ........((((.((((.((.(((((((..((.((.(((((..(..((((.......))))..)..)))------)).)))))))))))..((((........)))))).)))).)))). ( -57.90) >DroPse_CAF1 143120 108 + 1 UGAUGAUGAUGGGCGCCGGCCGCCGCCGCUGCUGCAGCGGCCAUCG---AGUAGCGGUCAUUGGCAGAG---------CCGCCACCCGUUUGCGGAAGGAGGCCCAGGCCAGCGGCGGCG ...........(((....)))((((((((((((((..((.....))---.)))))((((..(((....(---------((.((..(((....)))..)).))))))))))))))))))). ( -51.70) >DroGri_CAF1 182606 120 + 1 UGAUGAUGAUGGGCGCCGGCCGCUGCUGCCGCUGCCGCUGCCAUCGAUGAGUAACGGUCAUUGGCAGCGGCAGCUGCUGCGCCCGCAGCAUUUGGCAGCAGACCCAAGCCGGCAGCGGCG ............((((((((..(((((((((((((((((((((..(((........)))..)))))))))))))((((((....))))))...))))))))......)))))).)).... ( -73.40) >DroWil_CAF1 217179 108 + 1 UGAUGAUGAUGGGCUCCUGCCGCUGCGGCUGCUGCCGCGGCCAUUG---AAUAACGAUCGUUGGCAGCG---------GCUCCAGCUGUAUUUGGCAAAAGACCUAAGCCGGCGGCAGCG ................(((((((((((((((..(((((.((((.((---(.......))).)))).)))---------))..)))))))....(((...........))))))))))).. ( -49.80) >DroAna_CAF1 140117 108 + 1 UGGUGAUGAUGCGCUCCCGCUGCCGCCGCUGCUGCUGCGGCCAUGG---AAUACCGAUCAUUGGCAGCU---------GCGCCAGCUGCAUUUGGGAGCAGGCCCAAGCCGGCGGCUGCG ..........((((....(((((((((((.......)))))..(((---....)))......)))))).---------))))(((((((.((((((......))))))...))))))).. ( -48.30) >consensus UGAUGAUGAUGGGCGCCCGCCGCUGCCGCUGCUGCUGCGGCCAUCG___AAUAACGAUCAUUGGCAGCG_________GCGCCAGCAGCAUUUGGCAACAGACCCAAGCCGGCGGCAGCG ((.(((((((.(((....)))(((((.((....)).)))))...............))))))).)).............(((.(((.((.(((((........)))))...)).)))))) (-26.19 = -26.50 + 0.31)

| Location | 7,107,650 – 7,107,758 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -53.48 |
| Consensus MFE | -24.03 |
| Energy contribution | -25.32 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
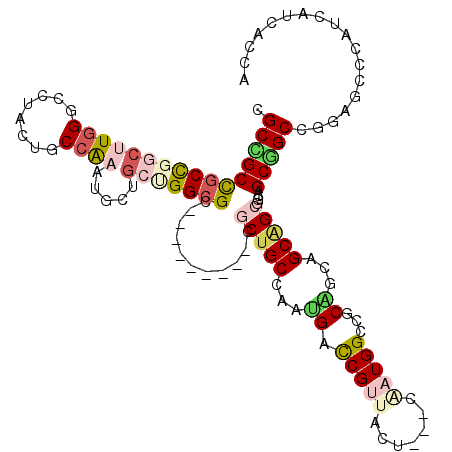
>2R_DroMel_CAF1 7107650 108 - 20766785 CGCUGCUGCCGGAUUGGGUUUAUUGCCACAAGCUGGAGGCGC---------CUCUGCCAAUGACCGUUAUU---CCAUGGCAGCAGCAGCGGCGGCUGCGGCGGGAGCCCAUCACCACCA .(((((((((((.((((((.....))).))).)))(((....---------)))(((((.((.........---.)))))))))))))))((.((.((.(((....)))))...)).)). ( -44.80) >DroVir_CAF1 174016 114 - 1 CGCGGCCGCCGGCUUGGGCCUAUUACCAAAUGCUGCCGGCGCAGC------GGCUGCCAAUGAUCGUUACUCAUCAAUGGCCGCAGCAGCUGCGGCAGCUGCUGGCGCCCACCAUCACCA ...((.(((((((((((........))))..((((((...(((((------.((.((((.((((........)))).)))).))....))))))))))).))))))).)).......... ( -53.40) >DroPse_CAF1 143120 108 - 1 CGCCGCCGCUGGCCUGGGCCUCCUUCCGCAAACGGGUGGCGG---------CUCUGCCAAUGACCGCUACU---CGAUGGCCGCUGCAGCAGCGGCGGCGGCCGGCGCCCAUCAUCAUCA ((((((((((((((.(((......))).....((((((((((---------.((.......))))))))))---))..))))(((((....)))))))))).)))))............. ( -55.30) >DroGri_CAF1 182606 120 - 1 CGCCGCUGCCGGCUUGGGUCUGCUGCCAAAUGCUGCGGGCGCAGCAGCUGCCGCUGCCAAUGACCGUUACUCAUCGAUGGCAGCGGCAGCGGCAGCAGCGGCCGGCGCCCAUCAUCAUCA ....((.(((((((...(.((((((((...((((((....))))))(((((((((((((((((.......))))...)))))))))))))))))))))))))))))))............ ( -73.60) >DroWil_CAF1 217179 108 - 1 CGCUGCCGCCGGCUUAGGUCUUUUGCCAAAUACAGCUGGAGC---------CGCUGCCAACGAUCGUUAUU---CAAUGGCCGCGGCAGCAGCCGCAGCGGCAGGAGCCCAUCAUCAUCA ..((((((((((((..((.(((..((........))..))))---------)((((((..((..((((...---.))))..)).)))))))))))..)))))))................ ( -45.10) >DroAna_CAF1 140117 108 - 1 CGCAGCCGCCGGCUUGGGCCUGCUCCCAAAUGCAGCUGGCGC---------AGCUGCCAAUGAUCGGUAUU---CCAUGGCCGCAGCAGCAGCGGCGGCAGCGGGAGCGCAUCAUCACCA .(((((((((((((((((......)))).....)))))))).---------.)))))...((((..(..((---((...(((((.((....)).)))))....))))..)...))))... ( -48.70) >consensus CGCCGCCGCCGGCUUGGGCCUACUGCCAAAUGCUGCUGGCGC_________CGCUGCCAAUGACCGUUACU___CAAUGGCCGCAGCAGCAGCGGCAGCGGCCGGAGCCCAUCAUCACCA .((((((((((((((((........)))).....)))))))...........(((((...((.(((((.......)))))...))...)))))....))))).................. (-24.03 = -25.32 + 1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:30 2006