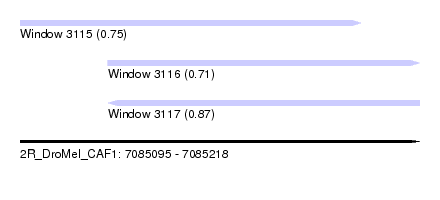
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,085,095 – 7,085,218 |
| Length | 123 |
| Max. P | 0.866171 |

| Location | 7,085,095 – 7,085,200 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.81 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -26.77 |
| Energy contribution | -27.77 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7085095 105 + 20766785 AGGCGACAUUUGUUAAUGUUCGGU-UUU------------UGCCGGC--AUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAGUUAACAUGGCAAG .((((((....))).(((((((((-(..------------((((...--..))))((((....))))...((((.....)))))))))))))).)))...((((((.......)))))). ( -31.20) >DroPse_CAF1 119472 107 + 1 AGGCGACAUUUGUUAAUGUUCGGC-AUU------------UGCCGGUAUAUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAAUGAACAUGGCAAC .((((((....))).((((((((.-...------------(((((.....)))))((((....))))...((((.....))))..)))))))).)))...((((((.......)))))). ( -29.50) >DroEre_CAF1 117871 117 + 1 AGGCGACAUUUGUUAAUGUUCGGU-UUUUAUUUUUUUUCUUGCCGGC--AUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAGUUAACAUGGCAAG .((((((....))).(((((((((-(...((((((((...((((...--..))))((((....)))).......)))))))).)))))))))).)))...((((((.......)))))). ( -34.90) >DroWil_CAF1 178459 103 + 1 AGGCGACAUUUGUUAAUGUUCGGU-UUU------------UGGC--U--CUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCAUGCGAACAUGGCAAU ..((..((.(((...(((((((((-(..------------...(--(--((....((((....)))).........))))...))))))))))....))).))((((....))))))... ( -27.32) >DroAna_CAF1 116786 106 + 1 AGGCGACAUUUGUUAAUGUUCGGUAUUU------------UGCCGGC--AUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAAUGAACAUGGCAAG .((((((....))).(((((((((....------------((((...--..))))((((....))))...((((.....)))).))))))))).)))...((((((.......)))))). ( -30.20) >DroPer_CAF1 117193 107 + 1 AGGCGACAUUUGUUAAUGUUCGGC-AUU------------UGCCGGUAUAUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAAUGAACAUGGCAAC .((((((....))).((((((((.-...------------(((((.....)))))((((....))))...((((.....))))..)))))))).)))...((((((.......)))))). ( -29.50) >consensus AGGCGACAUUUGUUAAUGUUCGGU_UUU____________UGCCGGC__AUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAAUGAACAUGGCAAG .((((((....))).(((((((((................(((((.....)))))((((....))))...((((.....)))).))))))))).)))...((((((.......)))))). (-26.77 = -27.77 + 1.00)

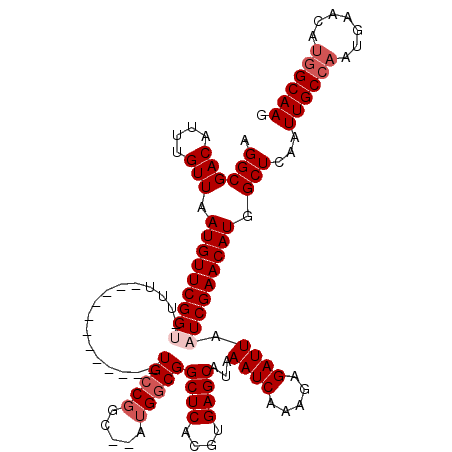
| Location | 7,085,122 – 7,085,218 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -20.25 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7085122 96 + 20766785 UGCCGGC--AUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAGUUAACAUGGCAAGCAG--CCC----------------CACCAACCAA-AA ....((.--.(((.(((((...(((((..........((......)).......))))).((((((.......))))))).))--)))----------------))))......-.. ( -25.43) >DroPse_CAF1 119499 116 + 1 UGCCGGUAUAUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAAUGAACAUGGCAACAAGACCAGGCGGAAGGAGGGAUUGCAGCCGCCAG-GA ..........((((((((..((((..(((.((((.....)))).........((((......)))))))..))))....(((..((...(....)..))..))).)))))))).-.. ( -34.00) >DroEre_CAF1 117910 98 + 1 UGCCGGC--AUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAGUUAACAUGGCAAGCUGACCCC----------------CACCGACCAA-AA ...(((.--.(((.((.(((.((((((..........((......)).......))))..((((((.......))))))))))).)))----------------))))).....-.. ( -25.03) >DroYak_CAF1 121802 96 + 1 UGCCGGU--AUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAGUUAACAUGGCAACCAAACUGC----------------CAACAA--AG-AA .(((...--..))).((((....))))...((((.....))))..((.(((.((((......)))))))....(((((.......)))----------------))....--.)-). ( -24.30) >DroAna_CAF1 116814 99 + 1 UGCCGGC--AUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAAUGAACAUGGCAAGCAAACCAA----------------AACCGACCAAGGA (((.(.(--(((.((((((....))))...((((.....))))...))..)))).)....((((((.......)))))))))......----------------..((......)). ( -23.70) >DroPer_CAF1 117220 116 + 1 UGCCGGUAUAUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAAUGAACAUGGCAACAAGACCAGGCGGAAGGAGGGAUUGCAGCCGCCAG-GA ..........((((((((..((((..(((.((((.....)))).........((((......)))))))..))))....(((..((...(....)..))..))).)))))))).-.. ( -34.00) >consensus UGCCGGC__AUGGCGGCUCACGUGAGCAUAAAUCAAAGAGAUUAAUCGAACAUGGCUCAAUUGCCAAUGAACAUGGCAACCAGACCAC________________CACCAACCAA_AA (((((.....(((((((((....))))..........(((.(((........))))))...))))).......)))))....................................... (-20.25 = -20.25 + 0.00)
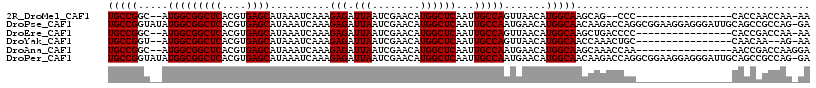
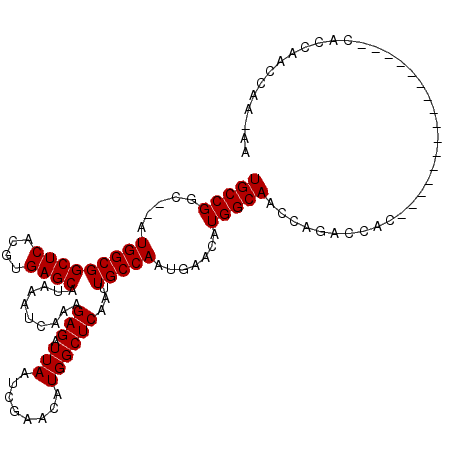
| Location | 7,085,122 – 7,085,218 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -22.15 |
| Energy contribution | -21.90 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7085122 96 - 20766785 UU-UUGGUUGGUG----------------GGG--CUGCUUGCCAUGUUAACUGGCAAUUGAGCCAUGUUCGAUUAAUCUCUUUGAUUUAUGCUCACGUGAGCCGCCAU--GCCGGCA ..-...((((((.----------------.((--(.((((((((.(....)))))...(((((.(((.((((.........))))..))))))))...)))).)))..--)))))). ( -32.30) >DroPse_CAF1 119499 116 - 1 UC-CUGGCGGCUGCAAUCCCUCCUUCCGCCUGGUCUUGUUGCCAUGUUCAUUGGCAAUUGAGCCAUGUUCGAUUAAUCUCUUUGAUUUAUGCUCACGUGAGCCGCCAUAUACCGGCA ..-.((((((((((((..((...........))..))))(((((.......)))))..(((((.(((.((((.........))))..))))))))....)))))))).......... ( -33.50) >DroEre_CAF1 117910 98 - 1 UU-UUGGUCGGUG----------------GGGGUCAGCUUGCCAUGUUAACUGGCAAUUGAGCCAUGUUCGAUUAAUCUCUUUGAUUUAUGCUCACGUGAGCCGCCAU--GCCGGCA ..-...(((((((----------------(.(((..((((((((.(....))))))).(((((.(((.((((.........))))..)))))))).))..))).))..--)))))). ( -33.80) >DroYak_CAF1 121802 96 - 1 UU-CU--UUGUUG----------------GCAGUUUGGUUGCCAUGUUAACUGGCAAUUGAGCCAUGUUCGAUUAAUCUCUUUGAUUUAUGCUCACGUGAGCCGCCAU--ACCGGCA ..-..--.....(----------------((.(((..(((((((.(....))))))))..)))(((((..(...((((.....))))....)..))))).)))(((..--...))). ( -30.70) >DroAna_CAF1 116814 99 - 1 UCCUUGGUCGGUU----------------UUGGUUUGCUUGCCAUGUUCAUUGGCAAUUGAGCCAUGUUCGAUUAAUCUCUUUGAUUUAUGCUCACGUGAGCCGCCAU--GCCGGCA ......((((((.----------------.(((((..(((((((.......)))))).(((((.(((.((((.........))))..)))))))).)..)))))....--)))))). ( -33.70) >DroPer_CAF1 117220 116 - 1 UC-CUGGCGGCUGCAAUCCCUCCUUCCGCCUGGUCUUGUUGCCAUGUUCAUUGGCAAUUGAGCCAUGUUCGAUUAAUCUCUUUGAUUUAUGCUCACGUGAGCCGCCAUAUACCGGCA ..-.((((((((((((..((...........))..))))(((((.......)))))..(((((.(((.((((.........))))..))))))))....)))))))).......... ( -33.50) >consensus UC_CUGGUCGGUG________________GUGGUCUGCUUGCCAUGUUAACUGGCAAUUGAGCCAUGUUCGAUUAAUCUCUUUGAUUUAUGCUCACGUGAGCCGCCAU__ACCGGCA .......................................((((..((....((((((((((((...))))))))((((.....))))...((((....)))).))))...)).)))) (-22.15 = -21.90 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:27 2006