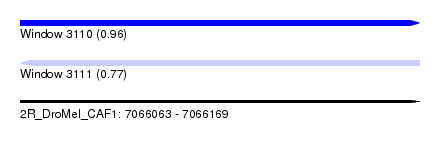
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,066,063 – 7,066,169 |
| Length | 106 |
| Max. P | 0.961354 |

| Location | 7,066,063 – 7,066,169 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -19.91 |
| Energy contribution | -21.47 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7066063 106 + 20766785 CAUCGGUUGUUGUCCGUUGUUAUUGUUGUUUCUGGUCGUCGGAUGUCGGACGUUGGACGUUGGACGUUGGACGUUGGACGUUGGACA-CUGGACAUUGGACGUUGGU ...........(((((.((...........((..((.(((.((((((.((((((.((((.....)))).)))))).)))))).))))-)..)))).)))))...... ( -40.30) >DroEre_CAF1 97099 85 + 1 CAGCGGUUGUUGUCCGUUGUUAUUGUUGUUUCUGGUCGUCGGAUGUCGGACGUUGGAC---------------------GCUGGACA-UUGGACAGUGGACGCUGGU (((((.(..(((((((.((((......((.((..(.((((.(....).)))))..)).---------------------))..))))-.)))))))..).))))).. ( -35.70) >DroYak_CAF1 98087 85 + 1 CAUCGGUUGUUGUCCGUUGUUAUUGUUGUUUCUGGUCGUCGGAUGUUGGACACUGGAC---------------------GUUGGACAU-UGGACACUGGACGUUGGU ((.(((.......))).))...........((..((.(((.((((((.(((.......---------------------))).)))))-).)))))..))....... ( -23.40) >consensus CAUCGGUUGUUGUCCGUUGUUAUUGUUGUUUCUGGUCGUCGGAUGUCGGACGUUGGAC_____________________GUUGGACA__UGGACACUGGACGUUGGU ...........(((((.((((..((((...((..(.((((.(....).)))))..))(((((((((.....)))))))))...))))....)))).)))))...... (-19.91 = -21.47 + 1.56)

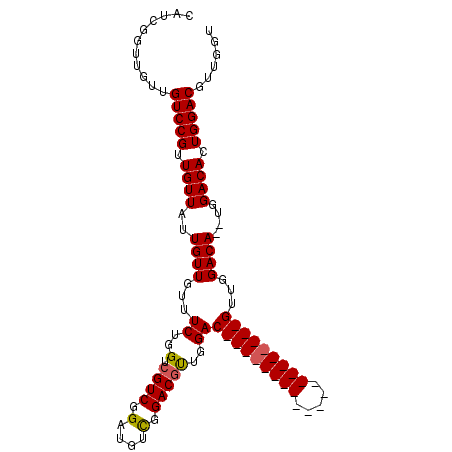
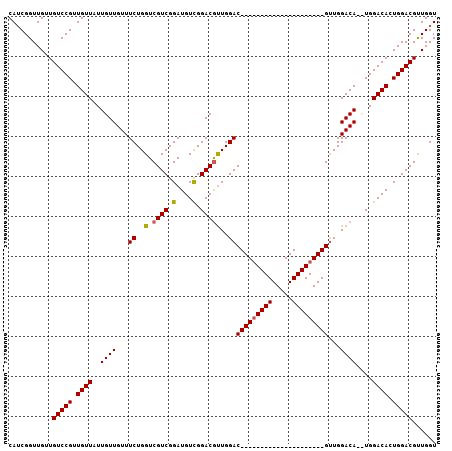
| Location | 7,066,063 – 7,066,169 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -18.37 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.30 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7066063 106 - 20766785 ACCAACGUCCAAUGUCCAG-UGUCCAACGUCCAACGUCCAACGUCCAACGUCCAACGUCCGACAUCCGACGACCAGAAACAACAAUAACAACGGACAACAACCGAUG ......((((..((((..(-((((..((((...((((..........))))...))))..)))))..))))....(......).........))))........... ( -19.90) >DroEre_CAF1 97099 85 - 1 ACCAGCGUCCACUGUCCAA-UGUCCAGC---------------------GUCCAACGUCCGACAUCCGACGACCAGAAACAACAAUAACAACGGACAACAACCGCUG ..(((((.....(((((.(-((((..((---------------------(.....)))..)))))....(.....)................))))).....))))) ( -19.80) >DroYak_CAF1 98087 85 - 1 ACCAACGUCCAGUGUCCA-AUGUCCAAC---------------------GUCCAGUGUCCAACAUCCGACGACCAGAAACAACAAUAACAACGGACAACAACCGAUG .....((((...(((...-.(((((..(---------------------(((..(((.....)))..))))....(......).........))))))))...)))) ( -15.40) >consensus ACCAACGUCCAAUGUCCA__UGUCCAAC_____________________GUCCAACGUCCGACAUCCGACGACCAGAAACAACAAUAACAACGGACAACAACCGAUG ......((((..((..(....)..)).............................((((........)))).....................))))........... (-10.52 = -10.30 + -0.22)

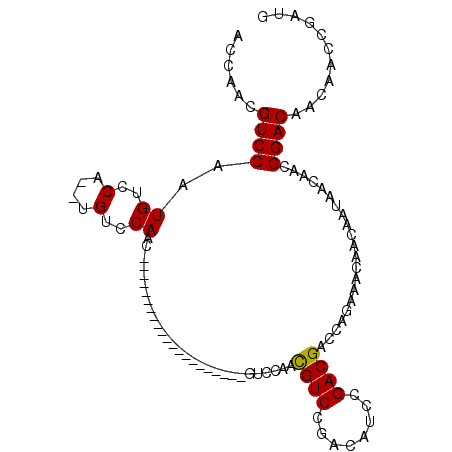
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:21 2006