
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,031,071 – 7,031,167 |
| Length | 96 |
| Max. P | 0.695755 |

| Location | 7,031,071 – 7,031,167 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.44 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -13.54 |
| Energy contribution | -14.65 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
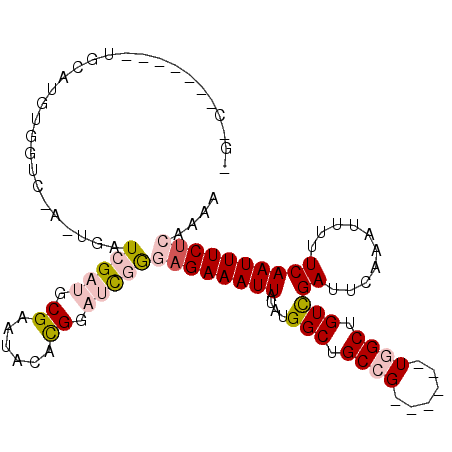
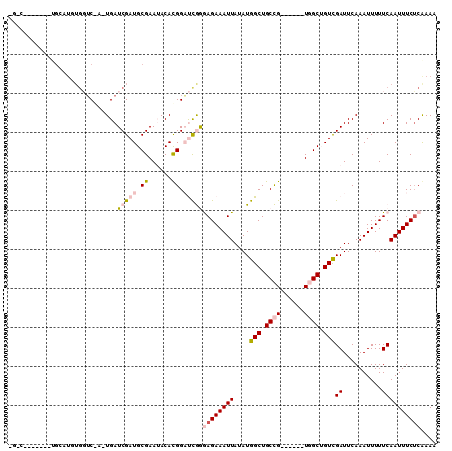
>2R_DroMel_CAF1 7031071 96 + 20766785 -UGU-------UGCAUUUGGUCGCUUGAUCGAUGCGAAUACACGGAUCGGGAGAAAUUAUAUGGCUGCCG------UGGCGGUCGAUUCAAAUUUUUCAAUUUCUCGAAA -..(-------((((((.((((....))))))))))).........((((((((........((((((..------..))))))((..........))..)))))))).. ( -27.70) >DroVir_CAF1 75448 101 + 1 ---AUGCGAUCAGCAUGUGUGC-UGUGAUCGAUGCGAAAACACGGCCUAAAAGAAAUUAUAUGGCUGCUGU-----UUGCUGUCGAUUCAAAUUUUUCAAUUUCUCGAAC ---...(((.(((((....)))-))((((((((((((..((((((((((.((....)).)).))))).)))-----)))).))))).)))..............)))... ( -27.10) >DroPse_CAF1 62377 103 + 1 UGACUACACUCUGAGUGUGGUCAA-UGAUCGAUGCGAAUACAUGGAUCGGGAGAAAUUAUAUGGCUGCCG------UGGCUGUCGAUUCAAAUUUUUCAAUUUCUCAAAA (((((((((.....))))))))).-(((((.(((......))).))))).((((((((....(((.((..------..)).)))((..........)))))))))).... ( -33.80) >DroYak_CAF1 60778 102 + 1 -GGU-------UGCAUUUGGUCGCUUGAUCGAUGCGAAUACACGGAUCGGGAGAAAUUAUAUGGCUGCCGUGGCCGUGGCGGUUGAUUCAAAUUUUUCAAUUUCUCAAAA -..(-------((((((.((((....)))))))))))...(((((..(((.((..((...))..)).)))...)))))(..(((((..........)))))..)...... ( -27.30) >DroMoj_CAF1 85329 85 + 1 -------------------UUC-AGUGAUCGAUGCGAAAACACGGCCCAAAAGAAAUUAUAUGGCUGCUGU-----UUGCUGUCGAUUCAAAUUUUUCAAUUUCACGAAC -------------------...-..((((((((((((..((((((((...((....))....))))).)))-----)))).))))).))).................... ( -18.70) >DroPer_CAF1 61263 103 + 1 UGACUACACUCUGAGUGUGGUCAA-UGAUCGAUGCGAAUACAUGGAUCGGGAGAAAUUAUAUGGCUGCCG------UGGCUGUCGAUUCAAAUUUUUCAAUUUCUCAAAA (((((((((.....))))))))).-(((((.(((......))).))))).((((((((....(((.((..------..)).)))((..........)))))))))).... ( -33.80) >consensus _G_C_______UGCAUGUGGUC_A_UGAUCGAUGCGAAUACACGGAUCGGGAGAAAUUAUAUGGCUGCCG______UGGCUGUCGAUUCAAAUUUUUCAAUUUCUCAAAA ............................(((((.((......)).)))))((((((((....(((.((((......)))).)))((..........)))))))))).... (-13.54 = -14.65 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:12 2006