
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,027,964 – 7,028,056 |
| Length | 92 |
| Max. P | 0.539989 |

| Location | 7,027,964 – 7,028,056 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 76.33 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -16.08 |
| Energy contribution | -15.75 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
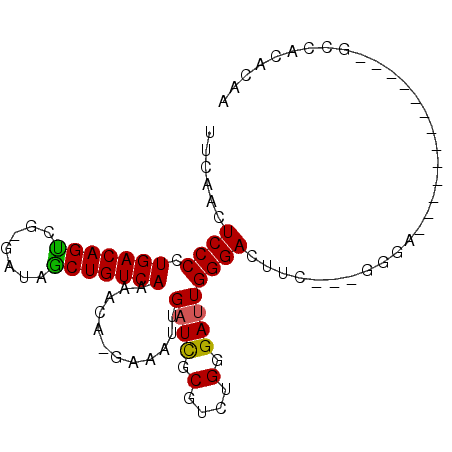
>2R_DroMel_CAF1 7027964 92 + 20766785 UUCAACUCCCCUGACAGUCG-GAUAGCUGUCAAAACA-GAAAUUGAUCGCGUCUGGGAUUGGGACUUC---GGGAAACCGAGCACAGAGCCACACAA .....(((((((((((((..-....))))))).....-......((((.(....).)))))))..(((---((....)))))....)))........ ( -26.00) >DroVir_CAF1 71906 83 + 1 UUCAAGUCCCCUGACAGUGG----AACUGUCAAAACA-GAAAUUGAUUGCGUCUGGGAAUGGGACUCAAAGGGGGACU--------G-UGCAUGCCA ....(((((((((((((...----..)))))......-........(((.((((.......)))).))).))))))))--------.-......... ( -26.60) >DroSec_CAF1 56309 90 + 1 UUCAACUCCCCUGACAGUCG-GAUAGCUGUCAAAACA-GAAAUUGAUCGCGUCUGGGAUUGGGACUUC---G-GACUUCG-GCACAGAGCCACACAA ..(((.((((..(((.((((-(....((((....)))-)...)))))...))).)))))))((.((..---(-..(....-)..)..))))...... ( -22.10) >DroEre_CAF1 58484 70 + 1 UUCAACUCCCCUGACAGCCG-GAUAGCUGUCAAAACA-GAAAUUGAUCGCGUCUGGGAUUGGGA-------------------------CCGCACAA ......((((.(((((((..-....))))))).....-......((((.(....).))))))))-------------------------........ ( -18.00) >DroYak_CAF1 57658 79 + 1 UUCAACUCCCCUGACAGUCG-CAUAGCUGUCAAAACA-GAAAUUGAUCGCGUCUGGGAUUGGGACUCC---GGGA-------------ACCACACAA ......((((....(((((.-((..((.(((((....-....))))).))...)).)))))((...))---))))-------------......... ( -19.20) >DroAna_CAF1 56010 82 + 1 UUCAACUCCCCUGACAGCCGGGAUAGCUGUCAAAACAUGAAAUUGAUCGCGUCUGGGAUUGGGACCU-----GCA-ACC---------CCUCUACUA ......((((.(((((((.......)))))))..((.(((......))).))..))))..(((....-----...-.))---------)........ ( -17.80) >consensus UUCAACUCCCCUGACAGUCG_GAUAGCUGUCAAAACA_GAAAUUGAUCGCGUCUGGGAUUGGGACUUC___GGGA_____________GCCACACAA ......((((.(((((((.......)))))))............((((.(....).))))))))................................. (-16.08 = -15.75 + -0.33)

| Location | 7,027,964 – 7,028,056 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 76.33 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -15.70 |
| Energy contribution | -16.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
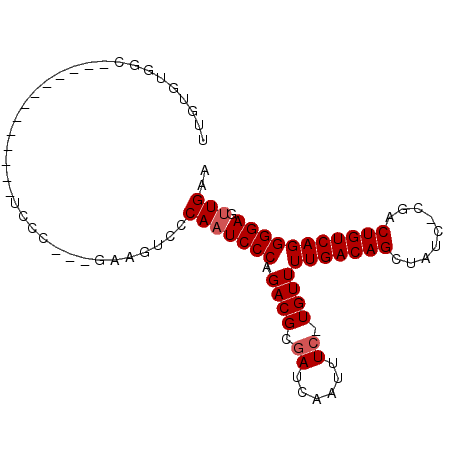
>2R_DroMel_CAF1 7027964 92 - 20766785 UUGUGUGGCUCUGUGCUCGGUUUCCC---GAAGUCCCAAUCCCAGACGCGAUCAAUUUC-UGUUUUGACAGCUAUC-CGACUGUCAGGGGAGUUGAA ..((.(((....(..(((((....))---).))..).....))).))....((((((((-(...(((((((.....-...)))))))))))))))). ( -26.20) >DroVir_CAF1 71906 83 - 1 UGGCAUGCA-C--------AGUCCCCCUUUGAGUCCCAUUCCCAGACGCAAUCAAUUUC-UGUUUUGACAGUU----CCACUGUCAGGGGACUUGAA .......((-.--------(((((((..(((.(((.........))).)))........-.....((((((..----...))))))))))))))).. ( -23.80) >DroSec_CAF1 56309 90 - 1 UUGUGUGGCUCUGUGC-CGAAGUC-C---GAAGUCCCAAUCCCAGACGCGAUCAAUUUC-UGUUUUGACAGCUAUC-CGACUGUCAGGGGAGUUGAA (((((((((.....))-)......-.---((........))....))))))((((((((-(...(((((((.....-...)))))))))))))))). ( -23.60) >DroEre_CAF1 58484 70 - 1 UUGUGCGG-------------------------UCCCAAUCCCAGACGCGAUCAAUUUC-UGUUUUGACAGCUAUC-CGGCUGUCAGGGGAGUUGAA ((((((((-------------------------........)).).)))))((((((((-(...(((((((((...-.)))))))))))))))))). ( -24.00) >DroYak_CAF1 57658 79 - 1 UUGUGUGGU-------------UCCC---GGAGUCCCAAUCCCAGACGCGAUCAAUUUC-UGUUUUGACAGCUAUG-CGACUGUCAGGGGAGUUGAA ((((((((.-------------..))---(((.......)))...))))))((((((((-(...(((((((.....-...)))))))))))))))). ( -21.90) >DroAna_CAF1 56010 82 - 1 UAGUAGAGG---------GGU-UGC-----AGGUCCCAAUCCCAGACGCGAUCAAUUUCAUGUUUUGACAGCUAUCCCGGCUGUCAGGGGAGUUGAA ..((...((---------(((-((.-----......)))))))..))....((((((((....((((((((((.....)))))))))))))))))). ( -28.40) >consensus UUGUGUGGC_____________UCCC___GAAGUCCCAAUCCCAGACGCGAUCAAUUUC_UGUUUUGACAGCUAUC_CGACUGUCAGGGGAGUUGAA ....................................(((((((.((((.((......)).))))(((((((.........))))))))))).))).. (-15.70 = -16.03 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:11 2006