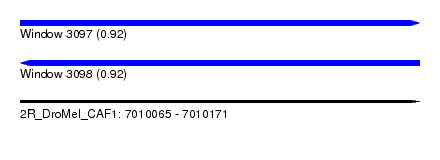
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,010,065 – 7,010,171 |
| Length | 106 |
| Max. P | 0.923654 |

| Location | 7,010,065 – 7,010,171 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.54 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -17.49 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
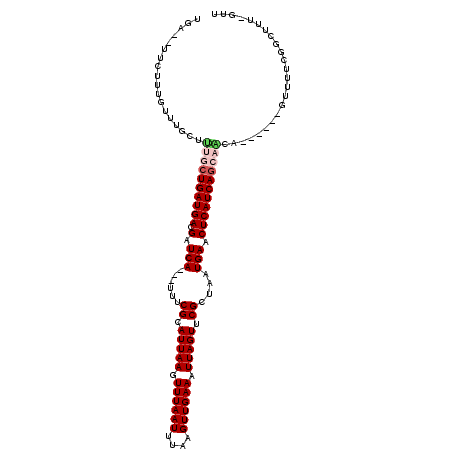
>2R_DroMel_CAF1 7010065 106 + 20766785 CGA--UUCUUUGUUUGCUUUGCUGAUGACGAUCA---UUUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGUUAAUGAACUCAUCAGCAACA------GUUUUUGACUUUUGUU ...--......(((.(((((((((((((.(.(((---((.((.(((((.((((((....)))))).))))).))..))))).))))))))))).)------))....)))....... ( -27.70) >DroVir_CAF1 51546 115 + 1 UGA--UUCUUUGUUUGCUUUGCUGAUGACGAUCAUCAUUUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAGCAGCAGCAGCAGUGUUCAGCCUUUGUU (((--....(((((.(((((((((((((.(.((((.....((.(((((.((((((....)))))).))))).))...)))).))))))))))).))))))))...)))......... ( -30.60) >DroWil_CAF1 56492 90 + 1 UGA--UUCUUUGUUUGCUUUACUGAUGACGAUCA---UUUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAGCU----------------------GGG ...--...........(((..(((((((.(.(((---((.((.(((((.((((((....)))))).))))).))..))))).))))))))..----------------------))) ( -19.90) >DroMoj_CAF1 55652 105 + 1 UGA--UUCUUUGUUUGCUCUGCUGAUGACGAUCAUCAGUUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAACAGCAGAC----UCUGCAGAC------ ...--......(((((((((((((.(((.((((((.((..((.(((((.((((((....)))))).))))).)))).))))..)).))).))))))).----...))))))------ ( -28.10) >DroAna_CAF1 39185 106 + 1 UAA--UUCUUUGUUUGCUUUGCUGAUGACGAUCA---UUUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAGGCACA------GUUUUGGGCUUAAGUU ...--..(((.(((((((.(((((((((.(.(((---((.((.(((((.((((((....)))))).))))).))..))))).))))))).))).)------))...))))..))).. ( -23.70) >DroPer_CAF1 40326 108 + 1 CUCUUCUCUUUGUUUGUUUUGCUGAUGAUGAUCA---UUUCGCAUUAAGUUUAAUUCAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAGCAACA------GUUUUUGGCUUUGGUU .............((((..(((((((((.(.(((---((.((.(((((.((((((....)))))).))))).))..))))).)))))))))))))------)............... ( -25.00) >consensus UGA__UUCUUUGUUUGCUUUGCUGAUGACGAUCA___UUUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAGCAACA______GUUUUCGGCUUU_GUU ..................((((((((((.(.(((......((.(((((.((((((....)))))).))))).))....))).)))))))))))........................ (-17.49 = -18.22 + 0.72)


| Location | 7,010,065 – 7,010,171 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.54 |
| Mean single sequence MFE | -20.99 |
| Consensus MFE | -15.73 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
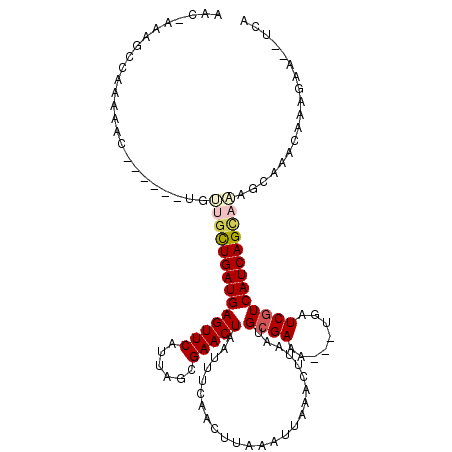
>2R_DroMel_CAF1 7010065 106 - 20766785 AACAAAAGUCAAAAAC------UGUUGCUGAUGAGUUCAUUAACGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAAA---UGAUCGUCAUCAGCAAAGCAAACAAAGAA--UCG ................------..((((((((((((((......))))).........................((((..---...))))))))))))).............--... ( -19.40) >DroVir_CAF1 51546 115 - 1 AACAAAGGCUGAACACUGCUGCUGCUGCUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAAAUGAUGAUCGUCAUCAGCAAAGCAAACAAAGAA--UCA ......(((........)))(((..((((((((((.((((((.((...((((((......)))))).........))...)))))).).))))))))).)))..........--... ( -26.00) >DroWil_CAF1 56492 90 - 1 CCC----------------------AGCUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAAA---UGAUCGUCAUCAGUAAAGCAAACAAAGAA--UCA ...----------------------.(((((((((.(((((.(((((.....))).......((((....))))))..))---))).).))))))))...............--... ( -16.10) >DroMoj_CAF1 55652 105 - 1 ------GUCUGCAGA----GUCUGCUGUUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAACUGAUGAUCGUCAUCAGCAGAGCAAACAAAGAA--UCA ------((.(((...----.(((((((.((((((..(((((((.(((.....))).......((((....)))).....))))))))))))).)))))))))).))......--... ( -27.40) >DroAna_CAF1 39185 106 - 1 AACUUAAGCCCAAAAC------UGUGCCUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAAA---UGAUCGUCAUCAGCAAAGCAAACAAAGAA--UUA ...............(------(.(((.(((((((.(((((.(((((.....))).......((((....))))))..))---))).).))))))))).))...........--... ( -16.10) >DroPer_CAF1 40326 108 - 1 AACCAAAGCCAAAAAC------UGUUGCUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUGAAUUAAACUUAAUGCGAAA---UGAUCAUCAUCAGCAAAACAAACAAAGAGAAGAG ................------..((((((((((..(((((.(((.......(((.....)))..........)))..))---)))...)))))))))).................. ( -20.93) >consensus AAC_AAAGCCAAAAAC______UGUUGCUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAAA___UGAUCGUCAUCAGCAAAGCAAACAAAGAA__UCA ........................((((((((((((((......))))).........................((((........))))))))))))).................. (-15.73 = -16.32 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:09 2006