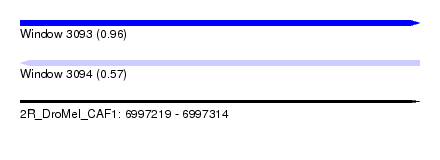
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,997,219 – 6,997,314 |
| Length | 95 |
| Max. P | 0.955502 |

| Location | 6,997,219 – 6,997,314 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.51 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -13.46 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6997219 95 + 20766785 AGAAAGCUCUAUUCGUGGA--UAGUGAGUGGUUAGAUAGG----------UGAAAACUAUUAAACGAGCGCACUUUAAACGCUCACUCAAUCACUUUUUGGUGCU-CC ....(((.(((...((((.--...(((((.(((.(((((.----------......))))).)))(((((.........)))))))))).))))....))).)))-.. ( -25.40) >DroSec_CAF1 25823 92 + 1 AGAAAACUCCAUCCGUGGA--U---GAGUGGUUAGAUAGG----------UGAAAACUAUUAAACGAGCGCACUUUAAGCGCUCACUCAAUCACUUUUCGGUGCU-AC .(((((.((((....))))--(---((((.(((.(((((.----------......))))).)))((((((.......))))))))))).....)))))......-.. ( -27.50) >DroSim_CAF1 24053 95 + 1 AGAAAGCUCUAUCCGUGGA--UAGUGAGUGGUUAGAUAGG----------UGAAAACUAUUAAACGAGCGCACUUUAAGCGCUCACUCAAUCACUUUUUGGUGCU-AC ....(((.(((...((((.--...(((((.(((.(((((.----------......))))).)))((((((.......))))))))))).))))....))).)))-.. ( -30.30) >DroEre_CAF1 28305 94 + 1 AGAAAGCUCUAUCCGCGGA--UAGUGAGUGCUUAGAUGGC----------UAAAACC-AUUAAACCAGCGCACUUUAAGCGCUCACUCAAUCACUUUCUGGUGCU-AC ....(((.(((......((--(((((((((((((((((((----------(......-........))).)).))))))))))))))..)))......))).)))-.. ( -29.04) >DroYak_CAF1 26266 95 + 1 AGAAAGCUCUAUCCGUGGA--UAGUGAGUGGUUAGAUGGG----------UAAGACCUAUUAAACGAGCGCACUUUAAGCGCUCACUCAAUCACUUGCCGGUGCU-GC ....(((.(((((....))--).(..(((((((.((((((----------.....))))))....((((((.......))))))....)))))))..).)).)))-.. ( -33.40) >DroAna_CAF1 28559 108 + 1 GGAAAGUUCUUGCCACUCACCUGGCGAGAGAUAUGAAGGUAGAAAAAAUAAGGAAACUAUUAAAUCAAUGGGCUUUAAGUGCACUCUCUAGCACUUUCUAUUGGAGGA ......(((((((((......))))))))).......((((((((...(((((...(((((.....))))).))))).((((........))))))))))))...... ( -27.50) >consensus AGAAAGCUCUAUCCGUGGA__UAGUGAGUGGUUAGAUAGG__________UGAAAACUAUUAAACGAGCGCACUUUAAGCGCUCACUCAAUCACUUUCUGGUGCU_AC .........................((((((((.(((((.................))))).))).(((((.......))))))))))...((((....))))..... (-13.46 = -13.88 + 0.42)


| Location | 6,997,219 – 6,997,314 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.51 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -11.81 |
| Energy contribution | -12.98 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6997219 95 - 20766785 GG-AGCACCAAAAAGUGAUUGAGUGAGCGUUUAAAGUGCGCUCGUUUAAUAGUUUUCA----------CCUAUCUAACCACUCACUA--UCCACGAAUAGAGCUUUCU ((-(((.............(((((((((((.......))))))(((..((((......----------.))))..))).)))))(((--(......)))).))))).. ( -21.70) >DroSec_CAF1 25823 92 - 1 GU-AGCACCGAAAAGUGAUUGAGUGAGCGCUUAAAGUGCGCUCGUUUAAUAGUUUUCA----------CCUAUCUAACCACUC---A--UCCACGGAUGGAGUUUUCU ..-(((.(((....(((..(((((((((((.......))))))(((..((((......----------.))))..))).))))---)--..)))...))).))).... ( -27.00) >DroSim_CAF1 24053 95 - 1 GU-AGCACCAAAAAGUGAUUGAGUGAGCGCUUAAAGUGCGCUCGUUUAAUAGUUUUCA----------CCUAUCUAACCACUCACUA--UCCACGGAUAGAGCUUUCU ..-(((.............(((((((((((.......))))))(((..((((......----------.))))..))).)))))(((--((....))))).))).... ( -27.10) >DroEre_CAF1 28305 94 - 1 GU-AGCACCAGAAAGUGAUUGAGUGAGCGCUUAAAGUGCGCUGGUUUAAU-GGUUUUA----------GCCAUCUAAGCACUCACUA--UCCGCGGAUAGAGCUUUCU ..-......(((((((...(((((.(((((.......))))).(((((((-(((....----------))))).))))))))))(((--((....))))).))))))) ( -33.40) >DroYak_CAF1 26266 95 - 1 GC-AGCACCGGCAAGUGAUUGAGUGAGCGCUUAAAGUGCGCUCGUUUAAUAGGUCUUA----------CCCAUCUAACCACUCACUA--UCCACGGAUAGAGCUUUCU ..-......(((.(((((((((((((((((.......))))))))))))).((.....----------.)).......))))..(((--((....))))).))).... ( -27.80) >DroAna_CAF1 28559 108 - 1 UCCUCCAAUAGAAAGUGCUAGAGAGUGCACUUAAAGCCCAUUGAUUUAAUAGUUUCCUUAUUUUUUCUACCUUCAUAUCUCUCGCCAGGUGAGUGGCAAGAACUUUCC ..........((((((.((.(((((.((.......)).....((...(((((.....)))))...))...........)))))((((......)))).)).)))))). ( -19.00) >consensus GU_AGCACCAAAAAGUGAUUGAGUGAGCGCUUAAAGUGCGCUCGUUUAAUAGUUUUCA__________CCCAUCUAACCACUCACUA__UCCACGGAUAGAGCUUUCU ...........(((((.(((((((((((((.......)))))))))))))......................((((.((...............)).))))))))).. (-11.81 = -12.98 + 1.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:05 2006