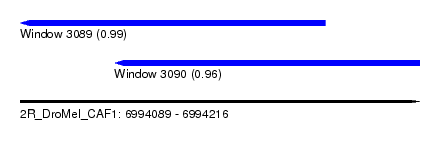
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,994,089 – 6,994,216 |
| Length | 127 |
| Max. P | 0.986337 |

| Location | 6,994,089 – 6,994,186 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.41 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -15.18 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
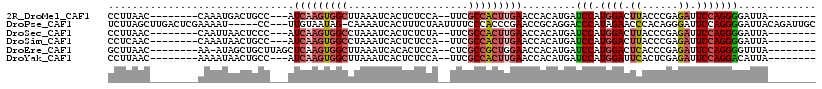
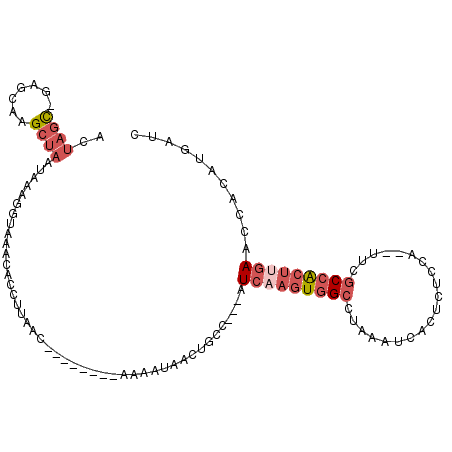
>2R_DroMel_CAF1 6994089 97 - 20766785 CCUUAAC--------CAAAUGACUGCC---AUCAAGUGGCUUAAAUCACUCUCCA--UUCGCCACUUGAACCACAUGAUCCAUGGACUUACCCGAGAUUCCAGGGGAUUA-------- .......--------............---.(((((((((...............--...)))))))))......((((((.(((((((....)))..))))..))))))-------- ( -23.67) >DroPse_CAF1 24213 109 - 1 UCUUAGCUUGACUCGAAAAU-----CC---UUGUAAUAG-CAAAAUCACUUUCUAAUUUUCCCACCCGAACCGCAGGACCCAUAGAACCCACAGGGAUUCCAGGGGAUUACAGAUUGC .....(((..((..(.....-----.)---..))...))-).......(..(((....(((((..((........)).......((((((...))).)))..)))))....)))..). ( -17.20) >DroSec_CAF1 22648 97 - 1 CCUUAAC--------CAAUUAACUCCC---AUCAAGUGGCCUAAAUCACUCUCUA--UUCGCCACUUGAACCACAUGAUCCAUGGACUUACCCGAGAUUCCAGGGGAUUA-------- .......--------............---.(((((((((...............--...)))))))))......((((((.(((((((....)))..))))..))))))-------- ( -23.17) >DroSim_CAF1 20909 97 - 1 CCUCAAC--------CAAAUAACUGCC---AUCAAGUGGCCUAAAUCACUCUCCA--UUCGCCACUUGAACCACAUGAUCCAUGGACUUACCCGAGAUUCCAGGGGAUUA-------- .......--------............---.(((((((((...............--...)))))))))......((((((.(((((((....)))..))))..))))))-------- ( -23.17) >DroEre_CAF1 25331 99 - 1 GCUUAAC--------AA-AUAGCUGCUUAGCUCAAGUGGCUUAAAUCACACUCCA--CUCGCCGCUGGAACCACAUGAUCCAUGGACUCACCCGAGAUUCCAGGGGUUUA-------- .......--------..-..(((..(((.....)))..)))..............--...(((.((((((...((((...))))..(((....))).)))))).)))...-------- ( -24.70) >DroYak_CAF1 23196 97 - 1 CCUUAAC--------AAAAUAACUGCC---AUCAAGUGGCUUAAAUCACUCUCCA--UUCGCCACUUGAACCACAUGAUCCAUGGAUUCACUCGAGAUUCCAGGACAUUA-------- .......--------............---.(((((((((...............--...))))))))).....(((.(((.((((.((......)).))))))))))..-------- ( -23.97) >consensus CCUUAAC________AAAAUAACUGCC___AUCAAGUGGCCUAAAUCACUCUCCA__UUCGCCACUUGAACCACAUGAUCCAUGGACUCACCCGAGAUUCCAGGGGAUUA________ ...............................(((((((((....................))))))))).........(((.((((.((......)).)))))))............. (-15.18 = -16.02 + 0.84)
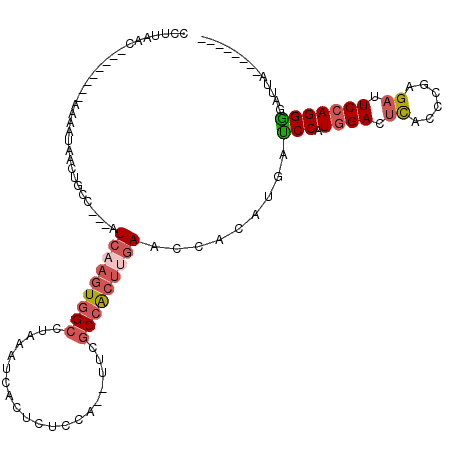
| Location | 6,994,119 – 6,994,216 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.70 |
| Mean single sequence MFE | -17.86 |
| Consensus MFE | -8.84 |
| Energy contribution | -10.23 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6994119 97 - 20766785 ACUAGCUGAGCAAGCUAAUAAAGGUAAACACCUUAAC--------CAAAUGACUGCC---AUCAAGUGGCUUAAAUCACUCUCCA--UUCGCCACUUGAACCACAUGAUC ..(((((.....)))))...(((((....)))))...--------............---.(((((((((...............--...)))))))))........... ( -22.17) >DroSec_CAF1 22678 97 - 1 ACUAGCAGAGCAAGCUAAUAAAGGUAAACACCUUAAC--------CAAUUAACUCCC---AUCAAGUGGCCUAAAUCACUCUCUA--UUCGCCACUUGAACCACAUGAUC ..((((.......))))...(((((....)))))...--------............---.(((((((((...............--...)))))))))........... ( -19.37) >DroSim_CAF1 20939 97 - 1 ACUAGCUGAGCAAGCUAAUAAAGGUAAACACCUCAAC--------CAAAUAACUGCC---AUCAAGUGGCCUAAAUCACUCUCCA--UUCGCCACUUGAACCACAUGAUC ..(((((.....)))))....((((....))))....--------............---.(((((((((...............--...)))))))))........... ( -20.17) >DroEre_CAF1 25361 98 - 1 ACUAGC-UAACAAGCUAAUAAAAGUAAACAGCUUAAC--------AA-AUAGCUGCUUAGCUCAAGUGGCUUAAAUCACACUCCA--CUCGCCGCUGGAACCACAUGAUC ....((-(((..(((((....((((.....))))...--------..-.)))))..)))))((.((((((...............--...)))))).))........... ( -19.77) >DroYak_CAF1 23226 97 - 1 ACUAGCACAGCAAGCUAAUAAAGCUAAACACCUUAAC--------AAAAUAACUGCC---AUCAAGUGGCUUAAAUCACUCUCCA--UUCGCCACUUGAACCACAUGAUC ............((((.....))))............--------............---.(((((((((...............--...)))))))))........... ( -16.37) >DroPer_CAF1 24243 97 - 1 ACCUUU-CAAGGAGCUAAUCAA---AAACGUCUUUGCUUGACUCGAAAAU-----CC---UUGUAAUAG-CAAAAUCACUUUCUAAUUUUCCCACCCGAACCGCAGGAUC .(((..-..)))..........---....(..((((((..((..(.....-----.)---..))...))-))))..)..................((........))... ( -9.30) >consensus ACUAGC_GAGCAAGCUAAUAAAGGUAAACACCUUAAC________AAAAUAACUGCC___AUCAAGUGGCCUAAAUCACUCUCCA__UUCGCCACUUGAACCACAUGAUC ..((((.......))))............................................(((((((((....................)))))))))........... ( -8.84 = -10.23 + 1.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:01 2006