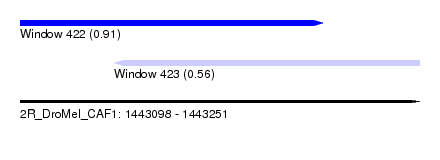
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,443,098 – 1,443,251 |
| Length | 153 |
| Max. P | 0.914737 |

| Location | 1,443,098 – 1,443,214 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.06 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -26.22 |
| Energy contribution | -26.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
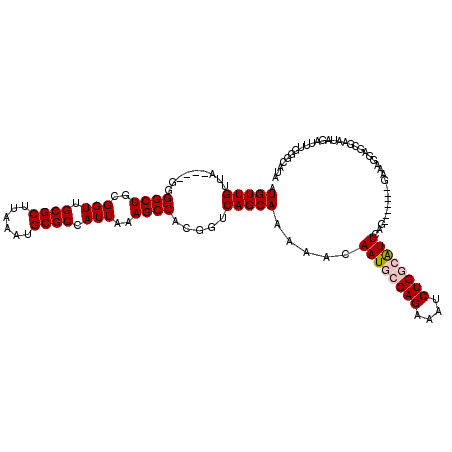
>2R_DroMel_CAF1 1443098 116 + 20766785 AUGUUGUUA----GGGGCUGCGGUUGCGCUUAAAUGCGCCAUUAAAGCCACGGUCAGCAAAAACAAUGCCAGAAAUCUGGCAUUCAGGGGCAGGGAAAGGAGCGAAUAGAUUUGGGCGUA .(((((...----(.((((..(((.((((......)))).)))..)))).)...))))).....((((((((....)))))))).................(((..((....))..))). ( -33.50) >DroSec_CAF1 61088 109 + 1 AUGUUGUUA----GGGGCUGCGGUUGCGCUUAAAUGCGCCAUUAAAGCCACGGUCAGCAAAAACAAUGCCAGAAAUCUGGCAUUCAG-------GAAAGGAGCGAGUAGAUUUGGCCAUA .(((((...----(.((((..(((.((((......)))).)))..)))).)...))))).....((((((((....))))))))...-------....((..((((....)))).))... ( -34.80) >DroSim_CAF1 59839 109 + 1 AUGUUGUUA----GGGGCUGCGGUUGCGCUUAAAUGCGCCAUUAAAGCCACGGUCAGCAAAAACAAUGCCAGAAAUCUGGCAUUCAG-------GAAAGGAGCGAAUAGAUUUGGCCAUA .(((((...----(.((((..(((.((((......)))).)))..)))).)...))))).....((((((((....))))))))...-------....((..((((....)))).))... ( -35.10) >DroEre_CAF1 70264 108 + 1 AUGUUGUUA----GGGGCUGCGGUUGCGCUUAAAUGCGCCAUUAAAGCCACGGUCAGCAAGAACAAUGCCAGAAAUCUGU-GUUAGG-------GAAAAAGGAGAACAGAUUUGGGCAUA .(((((...----(.((((..(((.((((......)))).)))..)))).)...)))))......(((((..((((((((-......-------...........)))))))).))))). ( -35.83) >DroYak_CAF1 65954 111 + 1 AUGUUGUUAGGUAGGGGCUGCGGUUGCGCUUAAAUGCGCCAUUAAAGCCACGGUCAGCAAAAACAAUCCCAGAAAUCUGU-GUUAGG--------AAAAGAGAGAAUAGAUUUGGGCAUA ..((((((..((...(((((.((((((((......))))......)))).))))).))...))))))((((..(((((((-......--------..........))))))))))).... ( -29.49) >consensus AUGUUGUUA____GGGGCUGCGGUUGCGCUUAAAUGCGCCAUUAAAGCCACGGUCAGCAAAAACAAUGCCAGAAAUCUGGCAUUCAG_______GAAAGGAGCGAAUAGAUUUGGGCAUA .(((((.........((((..(((.((((......)))).)))..)))).....))))).....((((((((....)))))))).................................... (-26.22 = -26.78 + 0.56)

| Location | 1,443,134 – 1,443,251 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -18.04 |
| Energy contribution | -19.08 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
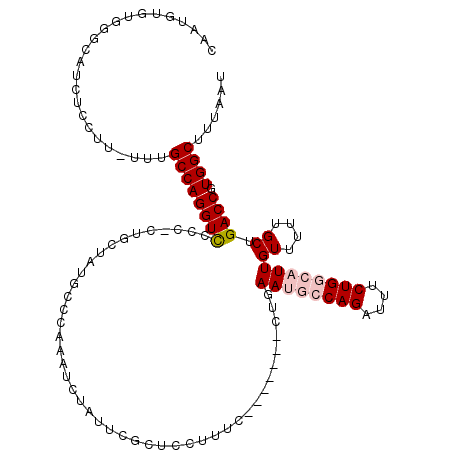
>2R_DroMel_CAF1 1443134 117 - 20766785 CAAUGUGUGGGCAUCUCCUUUUUUGCCAGGUCU---CUGCUACGCCCAAAUCUAUUCGCUCCUUUCCCUGCCCCUGAAUGCCAGAUUUCUGGCAUUGUUUUUGCUGACCGUGGCUUUAAU ........(((....)))......((((((((.---..((.((..............((..........)).....((((((((....))))))))))....)).)))).))))...... ( -27.70) >DroSec_CAF1 61124 112 - 1 CAAUGUGUGGGUAUCUCCUU-UUGGCCAGGUCCCCCCUGCUAUGGCCAAAUCUACUCGCUCCUUUC-------CUGAAUGCCAGAUUUCUGGCAUUGUUUUUGCUGACCGUGGCUUUAAU ......(((((((......(-((((((((((.......))).))))))))..))))))).......-------...((((((((....))))))))......(((......)))...... ( -32.80) >DroSim_CAF1 59875 113 - 1 CAAUGUGUGGGCAUCUCCUUUUUUGCCAGGUCCCCCCUGCUAUGGCCAAAUCUAUUCGCUCCUUUC-------CUGAAUGCCAGAUUUCUGGCAUUGUUUUUGCUGACCGUGGCUUUAAU .........((((..........))))(((.....)))(((((((.((..................-------...((((((((....))))))))((....)))).)))))))...... ( -28.70) >DroEre_CAF1 70300 108 - 1 CACUGUGUGGGCAUCACCUU-UUUGCCAGGUUCCU---CCUAUGCCCAAAUCUGUUCUCCUUUUUC-------CCUAAC-ACAGAUUUCUGGCAUUGUUCUUGCUGACCGUGGCUUUAAU (((.(.((.((((.......-......(((.....---)))(((((.((((((((...........-------......-))))))))..)))))......)))).))))))........ ( -22.83) >DroYak_CAF1 65994 104 - 1 CAAU-----GGCAUCUCCUG-GUCGCCAGGUCCCAAA-UCUAUGCCCAAAUCUAUUCUCUCUUUU--------CCUAAC-ACAGAUUUCUGGGAUUGUUUUUGCUGACCGUGGCUUUAAU ....-----(((........-)))((((((((.((((-......(((((((((............--------......-..)))))..))))......))))..)))).))))...... ( -22.55) >consensus CAAUGUGUGGGCAUCUCCUU_UUUGCCAGGUCCCC_CUGCUAUGCCCAAAUCUAUUCGCUCCUUUC_______CUGAAUGCCAGAUUUCUGGCAUUGUUUUUGCUGACCGUGGCUUUAAU ........................((((((((............................................((((((((....))))))))((....)).)))).))))...... (-18.04 = -19.08 + 1.04)

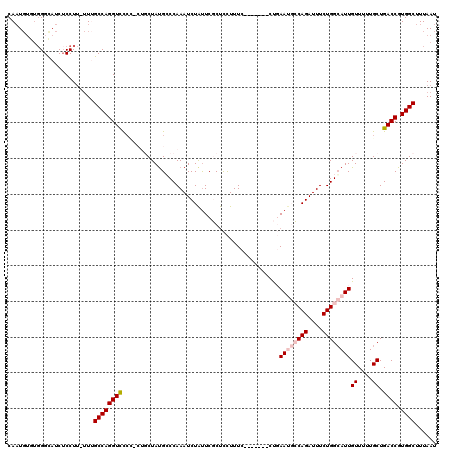
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:39 2006