| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
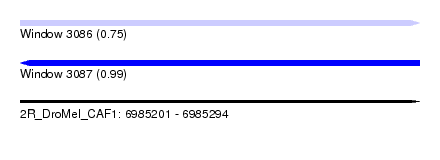
| Location | 6,985,201 – 6,985,294 |
| Length | 93 |
| Max. P | 0.985234 |

| Location | 6,985,201 – 6,985,294 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -13.58 |
| Energy contribution | -14.92 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
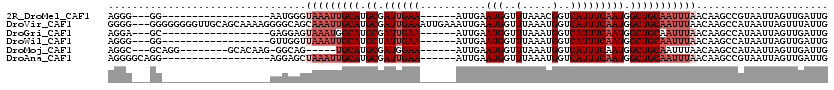
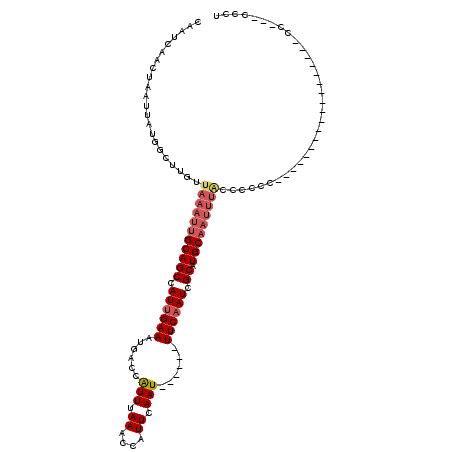
>2R_DroMel_CAF1 6985201 93 + 20766785 AGGG---GG------------------AAUGGGUAAAUUGCAUGCGAUUGAA------AUUGAAUGGUUUAAACGGUCAUUUCAAUGGCUGCAAUUUAACAAGCCGUAAUUAGUUGAUUG ....---((------------------..((..(((((((((.((.((((((------((.((.((.......)).)))))))))).))))))))))).))..))............... ( -26.90) >DroVir_CAF1 19034 117 + 1 GGGG---GGGGGGGGUUGCAGCAAAAGGGGCAGCAAAUUGCAUGCGAUUGAAAUUGAAAUUGAAUGGUUUAAAUGGUCAUUUCAAUGGCUGCAAUUUAACAAGCCAUAAUUAGUUUAUUG .((.---..(..((((((((((.......(((((.....)).))).((((((((.((.(((..........)))..)))))))))).))))))))))..)...))............... ( -29.30) >DroGri_CAF1 21544 93 + 1 AGGA---GC------------------GAGGAGUAAAUGGCAUGCGAUUGAA------AUUGAAUGGUUUAAAUGGUCAUUUCAAUGGCUGCAAUUUAACAAGCCAUAAUUAGUUGAUUG ..(.---((------------------..(...(((((.(((.((.((((((------((.((.(.((....)).))))))))))).))))).))))).)..)))............... ( -18.60) >DroWil_CAF1 23878 93 + 1 AGGG---GG------------------GUUGGUUAAAUUGCAUGCUAUUGAA------AUUGAAUGGUUUAAAUGGUCAUUUCAAUGGCUGCAAUUUAACAAGCCAUAAUUAGUUGAUUG ....---.(------------------(((.(((((((((((.(((((((((------((.((.(.((....)).))))))))))))))))))))))))).))))............... ( -31.80) >DroMoj_CAF1 22496 97 + 1 AGGC---GCAGG--------GCACAAG-GGCAG-----UGCAUGCGAUGGAA------AUUGAAUGGUUUAAAUGGUCAUUUCAAUGGCUGCAAUUUAACAAGCCAUAAUUAGUUGAUUG ...(---(((..--------((((...-....)-----))).)))).....(------((((((((((((((((((((((....))))))...))))))...)))))..))))))..... ( -22.90) >DroAna_CAF1 18936 96 + 1 AGGGGCAGG------------------AGGAGCUAAAUUGCAUGCGAUUGAA------AUUGAAUGGUUUAAAUGGUCAUUUCAAUGGCUGCAAUUUAACAAGCCGUAAUUAGUUGAUUG ...(((.(.------------------.....)(((((((((.((.((((((------((.((.(.((....)).))))))))))).)))))))))))....)))............... ( -23.10) >consensus AGGG___GG__________________GGGGAGUAAAUUGCAUGCGAUUGAA______AUUGAAUGGUUUAAAUGGUCAUUUCAAUGGCUGCAAUUUAACAAGCCAUAAUUAGUUGAUUG .................................(((((((((.((.((((((...........(((..(.....)..))))))))).)))))))))))...................... (-13.58 = -14.92 + 1.33)
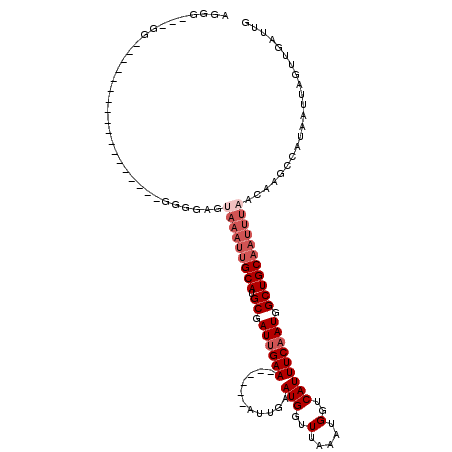
| Location | 6,985,201 – 6,985,294 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -20.08 |
| Consensus MFE | -11.08 |
| Energy contribution | -12.00 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.00 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6985201 93 - 20766785 CAAUCAACUAAUUACGGCUUGUUAAAUUGCAGCCAUUGAAAUGACCGUUUAAACCAUUCAAU------UUCAAUCGCAUGCAAUUUACCCAUU------------------CC---CCCU ...............((..((.(((((((((((.((((((((((..((....))...)).))------)))))).)).)))))))))..))..------------------))---.... ( -20.00) >DroVir_CAF1 19034 117 - 1 CAAUAAACUAAUUAUGGCUUGUUAAAUUGCAGCCAUUGAAAUGACCAUUUAAACCAUUCAAUUUCAAUUUCAAUCGCAUGCAAUUUGCUGCCCCUUUUGCUGCAACCCCCCCC---CCCC ...............(((..((.((((((((((.((((((((........................)))))))).)).)))))))))).))).....................---.... ( -21.36) >DroGri_CAF1 21544 93 - 1 CAAUCAACUAAUUAUGGCUUGUUAAAUUGCAGCCAUUGAAAUGACCAUUUAAACCAUUCAAU------UUCAAUCGCAUGCCAUUUACUCCUC------------------GC---UCCU .............(((((.(((......)))((.((((((((..................))------)))))).))..))))).........------------------..---.... ( -14.67) >DroWil_CAF1 23878 93 - 1 CAAUCAACUAAUUAUGGCUUGUUAAAUUGCAGCCAUUGAAAUGACCAUUUAAACCAUUCAAU------UUCAAUAGCAUGCAAUUUAACCAAC------------------CC---CCCU ...............((.(((((((((((((((.((((((((..................))------)))))).)).)))))))))).))).------------------))---.... ( -22.27) >DroMoj_CAF1 22496 97 - 1 CAAUCAACUAAUUAUGGCUUGUUAAAUUGCAGCCAUUGAAAUGACCAUUUAAACCAUUCAAU------UUCCAUCGCAUGCA-----CUGCC-CUUGUGC--------CCUGC---GCCU ...(((.......((((((.((......)))))))).....)))..................------......((((.(((-----(....-...))))--------..)))---)... ( -17.80) >DroAna_CAF1 18936 96 - 1 CAAUCAACUAAUUACGGCUUGUUAAAUUGCAGCCAUUGAAAUGACCAUUUAAACCAUUCAAU------UUCAAUCGCAUGCAAUUUAGCUCCU------------------CCUGCCCCU ...............(((..(((((((((((((.((((((((..................))------)))))).)).)))))))))))....------------------...)))... ( -24.37) >consensus CAAUCAACUAAUUAUGGCUUGUUAAAUUGCAGCCAUUGAAAUGACCAUUUAAACCAUUCAAU______UUCAAUCGCAUGCAAUUUACCCCCC__________________CC___CCCU ......................(((((((((((.((((((......(((.((....)).)))......)))))).)).)))))))))................................. (-11.08 = -12.00 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:58 2006