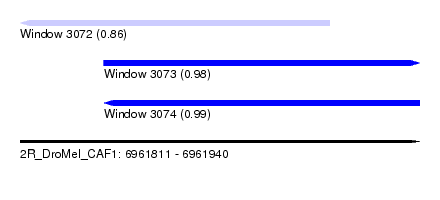
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,961,811 – 6,961,940 |
| Length | 129 |
| Max. P | 0.994585 |

| Location | 6,961,811 – 6,961,911 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 94.16 |
| Mean single sequence MFE | -27.31 |
| Consensus MFE | -25.40 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6961811 100 - 20766785 UUGAUCCCUUAGCCUGUUCCGAUUCCGCAUUCCCCCGAGCAAGCACAACUGUACCUGUCAUUGAGUUAUCAGCUGACUUUUGGGUAAGUUUGAGGAGAAA ....(((((((((.(((((.((........))....))))).))..((((.(((((((((((((....)))).))))....)))))))))))))).)).. ( -24.40) >DroSec_CAF1 5940 99 - 1 UUGAUCCCUCAGCUUGUUCCGAUCCCGCAUUCUCCCGAGCAAUCACAACUGUACCUGUCAUUGAGUUAUCAGCUGACUU-UGGGUAAGUUUUAGGAGAAA ..((((...(.....)....)))).....((((((.((....))..((((.(((((((((((((....)))).))))..-.)))))))))...)))))). ( -22.70) >DroSim_CAF1 13349 99 - 1 UUGAUCCCUCAGCUUGUUUCGAUCCCGCAUUCUCCCGAGCAAGCACAACUGUACCUGUCAUUGAGUUAUCAGCUGACUU-UGGGUAAGUUUGAGGAGAAA ....((((((((((((((.((..............)))))))))..((((.(((((((((((((....)))).))))..-.)))))))))))))).)).. ( -29.74) >DroEre_CAF1 12744 99 - 1 UUGAUCCCUCAGCUUGUUCCGAUUCCGCAUUUCCCCGAGCAAGCACAACUGUACCUGUCAUUGAGUUAUCAGCUGACUU-UGGGUAAGUUUGAGGGGAAA ....(((((((((((((((.((.........))...))))))))..((((.(((((((((((((....)))).))))..-.))))))))))))))))... ( -33.80) >DroYak_CAF1 6078 99 - 1 UUGAUCCCGCAGCUUGUUUCGAUUCCGCAUUCCCUCGAGCAAGCACAACUGUACCUGUCAUUGAGUUAUCGGCUGACUU-UGGGUAAGUUUGAGGAGAAA ....((((.(((((((((.(((............))))))))))..((((.(((((((((((((....)))).))))..-.))))))))))).)).)).. ( -25.90) >consensus UUGAUCCCUCAGCUUGUUCCGAUUCCGCAUUCCCCCGAGCAAGCACAACUGUACCUGUCAUUGAGUUAUCAGCUGACUU_UGGGUAAGUUUGAGGAGAAA ....(((((((((((((((.((........))....))))))))..((((.(((((((((((((....)))).))))....)))))))))))))).)).. (-25.40 = -25.52 + 0.12)

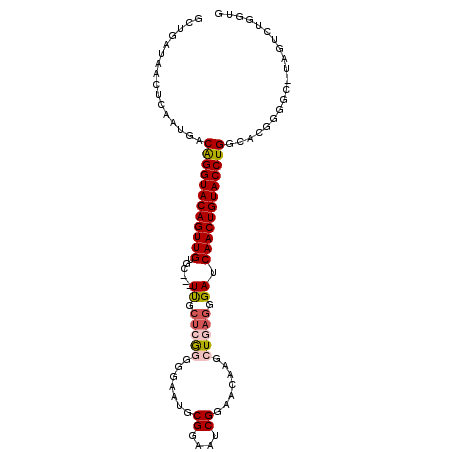
| Location | 6,961,838 – 6,961,940 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -20.44 |
| Energy contribution | -21.11 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
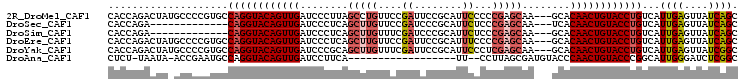
>2R_DroMel_CAF1 6961838 102 + 20766785 GCUGAUAACUCAAUGACAGGUACAGUUGUGC---UUGCUCGGGGGAAUGCGGAAUCGGAACAGGCUAAGGGAUCAACUGUACCUGGCACGGGGCAUAGUCUGGUG ................((((((((((((.((---(((.((.((...........)).)).)))))........)))))))))))).(((.((((...)))).))) ( -33.00) >DroSec_CAF1 5966 89 + 1 GCUGAUAACUCAAUGACAGGUACAGUUGUGA---UUGCUCGGGAGAAUGCGGGAUCGGAACAAGCUGAGGGAUCAACUGUACCUG-------------UCUGGUG .......(((....((((((((((((..(((---((.(((((.....(.((....)).).....))))).)))))))))))))))-------------)).))). ( -33.60) >DroSim_CAF1 13375 89 + 1 GCUGAUAACUCAAUGACAGGUACAGUUGUGC---UUGCUCGGGAGAAUGCGGGAUCGAAACAAGCUGAGGGAUCAACUGUACCUG-------------UCUGGUG .......(((....((((((((((((((...---((.(((((.......((....)).......))))).)).))))))))))))-------------)).))). ( -32.54) >DroEre_CAF1 12770 102 + 1 GCUGAUAACUCAAUGACAGGUACAGUUGUGC---UUGCUCGGGGAAAUGCGGAAUCGGAACAAGCUGAGGGAUCAACUGUACCUGGCACGGGGCAUAGUCUGGUG ................((((((((((((...---((.(((((.....(.((....)).).....))))).)).)))))))))))).(((.((((...)))).))) ( -33.70) >DroYak_CAF1 6104 102 + 1 GCCGAUAACUCAAUGACAGGUACAGUUGUGC---UUGCUCGAGGGAAUGCGGAAUCGAAACAAGCUGCGGGAUCAACUGUACCUGGCACGGGGCAUAGUCUGGUG ((((((..(((..((.((((((((((((.((---(((.((((............))))..)))))........)))))))))))).))..)))....))).))). ( -34.80) >DroAna_CAF1 15720 83 + 1 GCCGAGAUCCCAAUGCCGGGUACAGUUGGGUACAUCGCUAAGG--AA------------------UGAAGGAUCAACUGUACCUGGCAUUCGGU-UAUUA-AGAG (((((.......(((((((((((((((((...(.(((......--..------------------))).)..))))))))))))))))))))))-.....-.... ( -34.31) >consensus GCUGAUAACUCAAUGACAGGUACAGUUGUGC___UUGCUCGGGGGAAUGCGGAAUCGGAACAAGCUGAGGGAUCAACUGUACCUGGCACGGGGC_UAGUCUGGUG ................((((((((((((......((.(((((.......((....)).......))))).)).)))))))))))).................... (-20.44 = -21.11 + 0.67)
| Location | 6,961,838 – 6,961,940 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.27 |
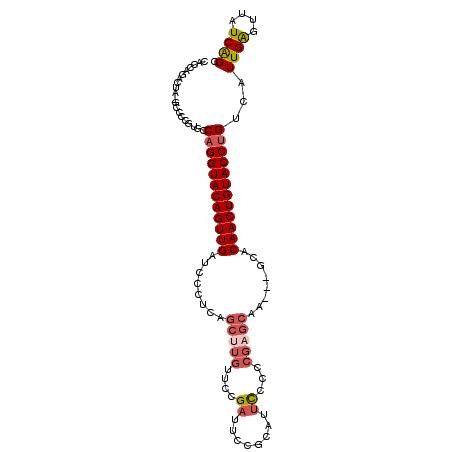
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6961838 102 - 20766785 CACCAGACUAUGCCCCGUGCCAGGUACAGUUGAUCCCUUAGCCUGUUCCGAUUCCGCAUUCCCCCGAGCAA---GCACAACUGUACCUGUCAUUGAGUUAUCAGC .....((....((.(.(((.(((((((((((((.....).((.(((((.((........))....))))).---)).)))))))))))).))).).))..))... ( -26.90) >DroSec_CAF1 5966 89 - 1 CACCAGA-------------CAGGUACAGUUGAUCCCUCAGCUUGUUCCGAUCCCGCAUUCUCCCGAGCAA---UCACAACUGUACCUGUCAUUGAGUUAUCAGC .....((-------------((((((((((((........(((((....((........))...)))))..---...)))))))))))))).((((....)))). ( -29.12) >DroSim_CAF1 13375 89 - 1 CACCAGA-------------CAGGUACAGUUGAUCCCUCAGCUUGUUUCGAUCCCGCAUUCUCCCGAGCAA---GCACAACUGUACCUGUCAUUGAGUUAUCAGC .....((-------------((((((((((((........(((((((.((..............)))))))---)).)))))))))))))).((((....)))). ( -31.84) >DroEre_CAF1 12770 102 - 1 CACCAGACUAUGCCCCGUGCCAGGUACAGUUGAUCCCUCAGCUUGUUCCGAUUCCGCAUUUCCCCGAGCAA---GCACAACUGUACCUGUCAUUGAGUUAUCAGC .....((....((.(.(((.((((((((((((........((((((((.((.........))...))))))---)).)))))))))))).))).).))..))... ( -30.40) >DroYak_CAF1 6104 102 - 1 CACCAGACUAUGCCCCGUGCCAGGUACAGUUGAUCCCGCAGCUUGUUUCGAUUCCGCAUUCCCUCGAGCAA---GCACAACUGUACCUGUCAUUGAGUUAUCGGC ...........(((..(((.((((((((((((........(((((((.(((............))))))))---)).)))))))))))).))).((....))))) ( -31.60) >DroAna_CAF1 15720 83 - 1 CUCU-UAAUA-ACCGAAUGCCAGGUACAGUUGAUCCUUCA------------------UU--CCUUAGCGAUGUACCCAACUGUACCCGGCAUUGGGAUCUCGGC ....-.....-.((.((((((.((((((((((......((------------------((--.......))))....)))))))))).)))))).))........ ( -30.60) >consensus CACCAGACUA_GCCCCGUGCCAGGUACAGUUGAUCCCUCAGCUUGUUCCGAUUCCGCAUUCCCCCGAGCAA___GCACAACUGUACCUGUCAUUGAGUUAUCAGC ....................((((((((((((........(((((....((........))...)))))........))))))))))))...((((....)))). (-19.82 = -20.72 + 0.91)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:46 2006