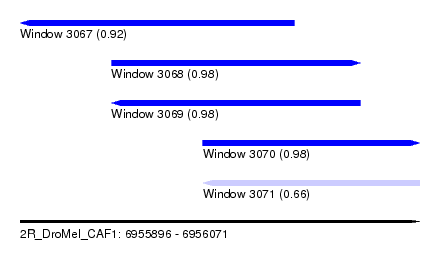
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,955,896 – 6,956,071 |
| Length | 175 |
| Max. P | 0.981527 |

| Location | 6,955,896 – 6,956,016 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -18.02 |
| Energy contribution | -17.83 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6955896 120 - 20766785 AUAGCCAGGCCAAAAGCCAAGGCCUUGAAGGUGCCAUCCUCAAUUUGGGUGCAUGCGCCGCAGGCAAAGAAAUUGAGGAUCAAAUUGCCAAGGAAUUCUCCAAUCAAGGCCUGCCACAAU ...((..(((.....))).(((((((((.(((...(((((((((((...(((.(((...))).)))...)))))))))))......)))..(((....)))..)))))))))))...... ( -44.70) >DroVir_CAF1 90 120 - 1 ACGCCCAGUCCAAAGGCCAACGCCUUAAAGGUGCCAUCCUCUGGCUGGGUGCAGGCGCCACAGGCGAAAAAGUUCAAUAUCAGAUUGCCAAUGAACUCGCCGAUGAGCGCUUGCCAAAGG ..((((((.(((.(((....((((.....))))....))).)))))))))((((((((((..(((((....(((((...............))))))))))..)).))))))))...... ( -50.86) >DroGri_CAF1 57 120 - 1 ACGCCCAGUCCAAAGGCGAGAGCCUUAAAGGUGCCAUCCUCCUGCUGGGUGCAGGCGCCACAGGCGAAAAAGUUCAAUAUCAAAUUGCCAAUGAAUUCGGCGAUGAGCGCCUGCCACAGA ..(((((((...(((((....)))))..(((......)))...)))))))((((((((((..(.((((...(((((((.....))))..)))...)))).)..)).))))))))...... ( -46.90) >DroWil_CAF1 57 120 - 1 ACACCCAGACCAAAGGCCAAGGCUUUGAAUGUGCCAUCUUCGAUUUGAGUGCAGGCGCCACAGGCAAAGAAAUUCAAUAUGAAAUUACCCAAAAAUUCACCAAUUAAAGCCUGCCAUAGA ..............(((..(((((((((.((((((.((........))(((....)))....))))......(((.....))).................)).))))))))))))..... ( -24.80) >DroMoj_CAF1 57 120 - 1 ACACCCAGGCCAAAGGCCAGCGCCUUGAAUGUGCCAUCCUCGGGCUGUGUGCAGGCGCCGCAGGCAAAGAAGUUCAAUAUCAGAUUGCCAAUGAACUCGCCGAUGAGCGCCUGCCAGAGA ...(((.((((...)))).((((.......)))).......)))((.((.((((((((((..(((...((.(((((...............))))))))))..)).)))))))))).)). ( -45.06) >DroAna_CAF1 21 120 - 1 AUGGCCAGGCCAAAAGCCAGAGCCUUGAAGGUACUAUCCUCCACUUGAGUGCAGGCACCGCAGGCAAAAAAGUUAAGAAUAAGAUUUCCCAGGAAUUCUCCAAUUAGGGCCUGCCAUAAU (((((.(((((....(((((.(((.....))).))..(((.(((....))).))).......))).......((((.....(((.(((....))).)))....))))))))))))))... ( -36.10) >consensus ACACCCAGGCCAAAGGCCAAAGCCUUGAAGGUGCCAUCCUCCAGCUGGGUGCAGGCGCCACAGGCAAAAAAGUUCAAUAUCAAAUUGCCAAUGAAUUCGCCAAUGAGCGCCUGCCACAGA .....((((...(((((....)))))..(((......)))...)))).(.(((((((((...))).....((((........))))......................)))))))..... (-18.02 = -17.83 + -0.19)

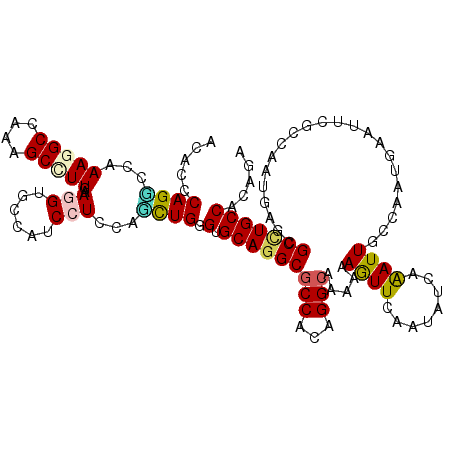
| Location | 6,955,936 – 6,956,045 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -41.12 |
| Consensus MFE | -25.32 |
| Energy contribution | -24.32 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6955936 109 + 20766785 AUCCUCAAUUUCUUUGCCUGCGGCGCAUGCACCCAAAUUGAGGAUGGCACCUUCAAGGCCUUGGCUUUUGGCCUGGCUAUCUUCAUGGCCAUCACGGUGGGUG----------GCUCGU (((((((((((...(((.(((...))).)))...)))))))))))(.(((((.(.(((((.........)))))((((((....))))))......).)))))----------.).... ( -42.50) >DroGri_CAF1 97 109 + 1 AUAUUGAACUUUUUCGCCUGUGGCGCCUGCACCCAGCAGGAGGAUGGCACCUUUAAGGCUCUCGCCUUUGGACUGGGCGUCUUCAUGGCCAUUACGGUGAGUG----------GAGUUG ......((((((((((((.(((((..((((.....))))(((((((.(.((...(((((....))))).))....).)))))))...)))))...)))))).)----------))))). ( -42.90) >DroEre_CAF1 7087 117 + 1 AUCCUCAAUUUCUUUGCCUGCGGUGCAUGCACCCAAAUCGAGGAUGGCACCUUCAAGGCCCUGGCUUUCGGCCUGGCCAUCUUCAUGGCCAUCACGGUGGGU--UCCCCAGGUGCUGUU ((((((.((((...(((.(((...))).)))...)))).))))))(((((((....(((((.(((.....)))(((((((....))))))).......))))--)....)))))))... ( -46.30) >DroYak_CAF1 65 111 + 1 AUCCUCAACUUCUUUGCCUGCGGUGCAUGCACCCAAGUUGAGGAUGGCACCUUCAAGGCCCUGGCCUUUGGCCUGGCCAUCUUCAUGGCCAUCACGGUGGGUGUUUCCAAU-------- (((((((((((...(((.(((...))).)))...)))))))))))(((((((..(((((....)))))..((((((((((....)))))))....))))))))))......-------- ( -47.20) >DroMoj_CAF1 97 108 + 1 AUAUUGAACUUCUUUGCCUGCGGCGCCUGCACACAGCCCGAGGAUGGCACAUUCAAGGCGCUGGCCUUUGGCCUGGGUGUCUUCAUGGCCAUUACGGUGAGCA----------AAGCU- ...........(((((((((.(((....)).).))).(((..((((((.(((..((((((((((((...))))..)))))))).))))))))).)))...)))----------)))..- ( -38.70) >DroAna_CAF1 61 109 + 1 AUUCUUAACUUUUUUGCCUGCGGUGCCUGCACUCAAGUGGAGGAUAGUACCUUCAAGGCUCUGGCUUUUGGCCUGGCCAUUUUCAUGGCUAUAACGGUAAGUU----------UUUUUU ............((((((...((.((((((......))(((((......))))).)))).))(((.....)))(((((((....)))))))....))))))..----------...... ( -29.10) >consensus AUCCUCAACUUCUUUGCCUGCGGCGCAUGCACCCAAAUCGAGGAUGGCACCUUCAAGGCCCUGGCCUUUGGCCUGGCCAUCUUCAUGGCCAUCACGGUGAGUG__________GAUCUU ...((((........(((((((.....))))........(((((((((....(((((((....))))).))....)))))))))..)))........)))).................. (-25.32 = -24.32 + -1.00)

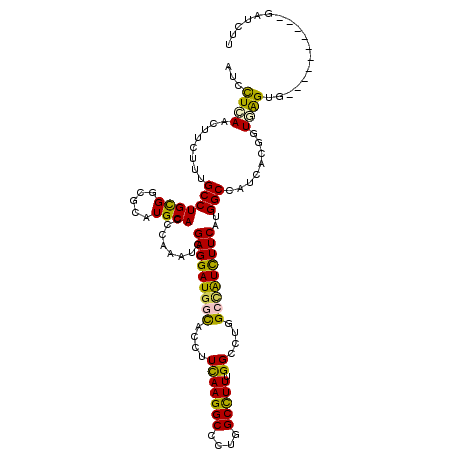
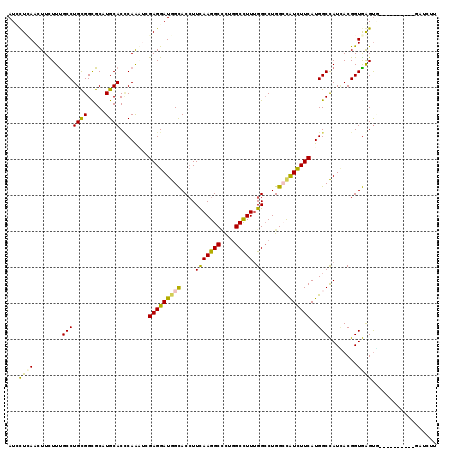
| Location | 6,955,936 – 6,956,045 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -24.27 |
| Energy contribution | -26.05 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6955936 109 - 20766785 ACGAGC----------CACCCACCGUGAUGGCCAUGAAGAUAGCCAGGCCAAAAGCCAAGGCCUUGAAGGUGCCAUCCUCAAUUUGGGUGCAUGCGCCGCAGGCAAAGAAAUUGAGGAU ......----------....((((.....((((.((........))(((.....)))..)))).....))))..(((((((((((...(((.(((...))).)))...))))))))))) ( -36.40) >DroGri_CAF1 97 109 - 1 CAACUC----------CACUCACCGUAAUGGCCAUGAAGACGCCCAGUCCAAAGGCGAGAGCCUUAAAGGUGCCAUCCUCCUGCUGGGUGCAGGCGCCACAGGCGAAAAAGUUCAAUAU .((((.----------...((.((....(((((.((....((((((((...(((((....)))))..(((......)))...)))))))))).).))))..)).))...))))...... ( -32.80) >DroEre_CAF1 7087 117 - 1 AACAGCACCUGGGGA--ACCCACCGUGAUGGCCAUGAAGAUGGCCAGGCCGAAAGCCAGGGCCUUGAAGGUGCCAUCCUCGAUUUGGGUGCAUGCACCGCAGGCAAAGAAAUUGAGGAU ....((((((.(((.--.(((.......(((((((....)))))))((.(....))).))))))...)))))).(((((((((((...(((.(((...))).)))...))))))))))) ( -51.60) >DroYak_CAF1 65 111 - 1 --------AUUGGAAACACCCACCGUGAUGGCCAUGAAGAUGGCCAGGCCAAAGGCCAGGGCCUUGAAGGUGCCAUCCUCAACUUGGGUGCAUGCACCGCAGGCAAAGAAGUUGAGGAU --------..((....))..((((((..(((((((....))))))).))..(((((....)))))...))))..(((((((((((...(((.(((...))).)))...))))))))))) ( -48.10) >DroMoj_CAF1 97 108 - 1 -AGCUU----------UGCUCACCGUAAUGGCCAUGAAGACACCCAGGCCAAAGGCCAGCGCCUUGAAUGUGCCAUCCUCGGGCUGUGUGCAGGCGCCGCAGGCAAAGAAGUUCAAUAU -..(((----------(((...(((..((((((((.(((.(.(...((((...)))).).).)))..))).)))))...))).(((((.((....)))))))))))))........... ( -35.80) >DroAna_CAF1 61 109 - 1 AAAAAA----------AACUUACCGUUAUAGCCAUGAAAAUGGCCAGGCCAAAAGCCAGAGCCUUGAAGGUACUAUCCUCCACUUGAGUGCAGGCACCGCAGGCAAAAAAGUUAAGAAU ......----------(((((.........(((((....)))))..(((.....)))...((((....(((.....(((.(((....))).))).)))..))))....)))))...... ( -27.90) >consensus AAAAGC__________CACCCACCGUGAUGGCCAUGAAGAUGGCCAGGCCAAAAGCCAGGGCCUUGAAGGUGCCAUCCUCAACUUGGGUGCAGGCACCGCAGGCAAAGAAGUUGAGGAU ............................(((((((....)))))))(((.....)))...((((....((((((((((.......)))))...)))))..))))............... (-24.27 = -26.05 + 1.78)
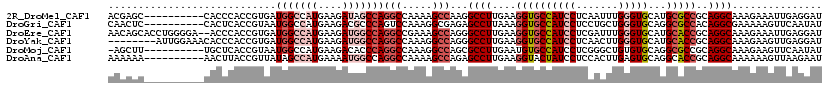
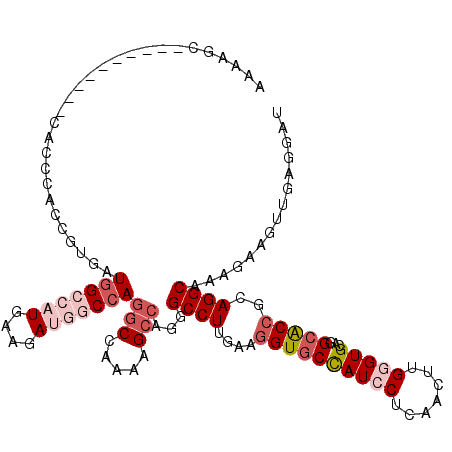

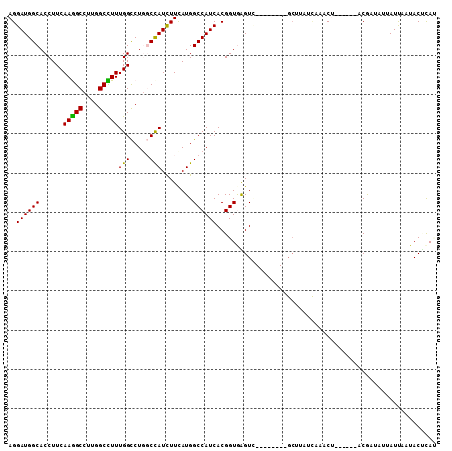
| Location | 6,955,976 – 6,956,071 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 75.56 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -19.18 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6955976 95 + 20766785 AGGAUGGCACCUUCAAGGCCUUGGCUUUUGGCCUGGCUAUCUUCAUGGCCAUCACGGUGGGUG--------GCUCGUUAAUCU------ACGACAUUAUUCAUACCCAU ..((((((........((((..(((.....))).)))).........))))))...(((((((--------..((((......------)))).........))))))) ( -31.03) >DroPse_CAF1 137 100 + 1 AGGAUGGCACCUUCAAGGCCUUGGCCUUUGGUCUCUCCAUCUUCAUGGCCAUCACGGUAAGUC--------GCAUAUCAAACAGAAGAUCCAAUCCAAUCAAUACUCA- .((((((...((((..((((.(((....(((.....)))...))).)))).....((((....--------...)))).....))))..)).))))............- ( -23.60) >DroSec_CAF1 101 95 + 1 AGGAUGGCACCUUCAAAGCCUUGGCUUUUGGCCUGGCCAUUUUCAUGGCCAUCACGGUGAGUU--------GCUUCUUAAUCU------ACGAAAUUAUUAAUACAUAU ((((.((((.(((....(((..(((.....)))(((((((....)))))))....)))))).)--------))))))).....------.................... ( -24.20) >DroSim_CAF1 7148 95 + 1 AGGAUGGCACCUUCAAAGCCUUGGCUUUUGGCCUGGCCAUCUUUAUGGCCAUCACGGUGGGUU--------GCUCCUCAACCU------ACGACAUUAUUCAUACGUAU ..((((((.........(((..(((.....))).)))..........))))))((((((((((--------(.....))))))------))((......))...))).. ( -31.71) >DroYak_CAF1 105 96 + 1 AGGAUGGCACCUUCAAGGCCCUGGCCUUUGGCCUGGCCAUCUUCAUGGCCAUCACGGUGGGUGUUUCCAAU-------AUACU------GCGAUGAUAUUCAUACUCAC ..((((((........((((..((((...)))).)))).........))))))...((((((((....(((-------((.(.------.....)))))).)))))))) ( -31.93) >DroPer_CAF1 137 100 + 1 AGGAUGGCACCUUCAAGGCCUUGGCCUUUGGUCUCUCCAUCUUCAUGGCCAUCACGGUAAGUC--------GCAUAUCAAACAGAAGAUCCAAUCCAAUCAAUACUCA- .((((((...((((..((((.(((....(((.....)))...))).)))).....((((....--------...)))).....))))..)).))))............- ( -23.60) >consensus AGGAUGGCACCUUCAAGGCCUUGGCCUUUGGCCUGGCCAUCUUCAUGGCCAUCACGGUGAGUC________GCUUAUCAAACU______ACGAUAUUAUUAAUACUCAU ..((((((......(((((....)))))(((.....)))........))))))........................................................ (-19.18 = -18.57 + -0.61)

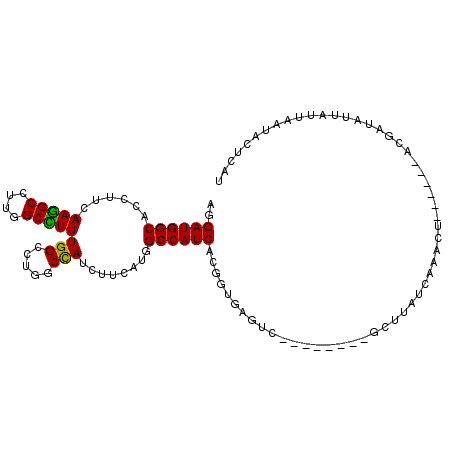
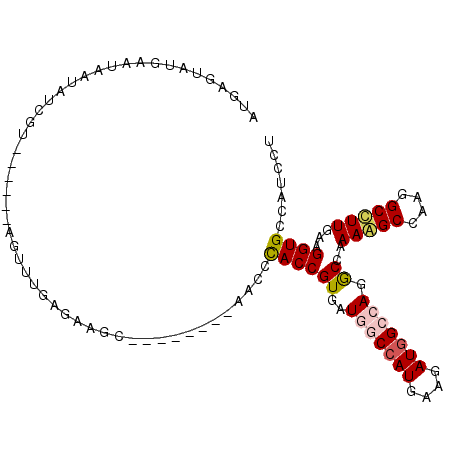
| Location | 6,955,976 – 6,956,071 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.56 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -18.73 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6955976 95 - 20766785 AUGGGUAUGAAUAAUGUCGU------AGAUUAACGAGC--------CACCCACCGUGAUGGCCAUGAAGAUAGCCAGGCCAAAAGCCAAGGCCUUGAAGGUGCCAUCCU .(((((((.....)).((((------......))))..--------.)))))..(.(((((((.(.(((...(((.(((.....)))..)))))).)..).))))))). ( -24.00) >DroPse_CAF1 137 100 - 1 -UGAGUAUUGAUUGGAUUGGAUCUUCUGUUUGAUAUGC--------GACUUACCGUGAUGGCCAUGAAGAUGGAGAGACCAAAGGCCAAGGCCUUGAAGGUGCCAUCCU -...((((..(..(((........)))..)..))))((--------(......)))(((((((((....))).....(((.(((((....)))))...))))))))).. ( -26.70) >DroSec_CAF1 101 95 - 1 AUAUGUAUUAAUAAUUUCGU------AGAUUAAGAAGC--------AACUCACCGUGAUGGCCAUGAAAAUGGCCAGGCCAAAAGCCAAGGCUUUGAAGGUGCCAUCCU ...(((.((..(((((....------.)))))..))))--------)...((((((..(((((((....))))))).))..(((((....)))))...))))....... ( -25.20) >DroSim_CAF1 7148 95 - 1 AUACGUAUGAAUAAUGUCGU------AGGUUGAGGAGC--------AACCCACCGUGAUGGCCAUAAAGAUGGCCAGGCCAAAAGCCAAGGCUUUGAAGGUGCCAUCCU .(((((((((......))))------)(((((.....)--------))))...)))).(((((((....)))))))((((.(((((....)))))....).)))..... ( -31.40) >DroYak_CAF1 105 96 - 1 GUGAGUAUGAAUAUCAUCGC------AGUAU-------AUUGGAAACACCCACCGUGAUGGCCAUGAAGAUGGCCAGGCCAAAGGCCAGGGCCUUGAAGGUGCCAUCCU .....((((....(((((((------.((..-------..((....))...)).))))))).))))..((((((...(((.(((((....)))))...))))))))).. ( -31.40) >DroPer_CAF1 137 100 - 1 -UGAGUAUUGAUUGGAUUGGAUCUUCUGUUUGAUAUGC--------GACUUACCGUGAUGGCCAUGAAGAUGGAGAGACCAAAGGCCAAGGCCUUGAAGGUGCCAUCCU -...((((..(..(((........)))..)..))))((--------(......)))(((((((((....))).....(((.(((((....)))))...))))))))).. ( -26.70) >consensus AUGAGUAUGAAUAAUAUCGU______AGUUUGAGAAGC________AACCCACCGUGAUGGCCAUGAAGAUGGCCAGGCCAAAAGCCAAGGCCUUGAAGGUGCCAUCCU ..................................................((((((..(((((((....))))))).))..(((((....)))))...))))....... (-18.73 = -18.95 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:43 2006