
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,905,631 – 6,905,726 |
| Length | 95 |
| Max. P | 0.816790 |

| Location | 6,905,631 – 6,905,726 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -13.52 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6905631 95 - 20766785 UUU-----CUCGGGGGAGGUU----CAUCAAUGGUAAUAGCAACUAUAAAUAGUUCCAGUAAAUUGGCCAUAACCAAAAGGCUGAACAAACAGCUCCCA----CCG----AA ...-----.(((((((((...----.....(((((.......))))).....((((.(((...((((......))))...)))))))......))))).----)))----). ( -27.50) >DroPse_CAF1 21837 94 - 1 ------------GGAGAGGCUGCCGUAGCAAUAAUACU-GUAUCUAUAAAUAGUGCCAGUAAAUUGGCCAUAAC-AAAUGGCUGAACAAAUAGUUUGCAUUGCCCA----AA ------------...(.((((((...(((.....((((-(...(((....)))...)))))..(..(((((...-..)))))..).......))).)))..)))).----.. ( -22.00) >DroSec_CAF1 21123 94 - 1 UU------CUCGGGGGAGUUU----UGUCAAUGGUAAUAGCAACUAUAAAUAGCUACAGCAAAUUGGCCAUAACCAAAUGGCUGAACAAACAGCUCCCA----CCG----AA ..------.((((((((((((----(((.....((..((((...........))))..))...(..(((((......)))))..))))))..)))))).----)))----). ( -31.70) >DroSim_CAF1 25186 95 - 1 UUG-----CUCGGGGUAGGUU----CAUCAAUGGUAAUAGCAACUAUAAAUAGUAUCAGUAAAUUGGCCAUAACCAAAUGGCUGAACAAACAGCUCCCA----CCG----AA ...-----.((((((.((...----.....(((((.......))))).....((.(((((...((((......))))...)))))))......)).)).----)))----). ( -20.70) >DroEre_CAF1 26050 95 - 1 UUU-----UUCGGGGGAGGCG----CAUCAAUGGUAAUAACAGCUAUAAAUAGUUCCAGUAAAUUGGCCAUAACCAAAAGGCUGAACAAAUAGCUCCCA----GCG----AA ...-----((((.(((((.(.----.....(((((.......))))).....((((.(((...((((......))))...))))))).....)))))).----.))----)) ( -24.40) >DroYak_CAF1 26724 104 - 1 UUUUUUUCUUCGGGGGAGGUG----CAUCAAUGGUAAUAGCAACUAUAAAUAGUUCCAGUAAAUUGGCCAUAACCAAAAGGCUGAACAAACAGCUCGCA----AGGCUUCAA .((((..(....)..))))..----.....(((((.......))))).....((((.(((...((((......))))...)))))))....(((.(...----.)))).... ( -21.40) >consensus UUU_____CUCGGGGGAGGUU____CAUCAAUGGUAAUAGCAACUAUAAAUAGUUCCAGUAAAUUGGCCAUAACCAAAAGGCUGAACAAACAGCUCCCA____CCG____AA .............(((((............(((((.......))))).....(((.((((...((((......))))...)))))))......))))).............. (-13.52 = -14.38 + 0.86)

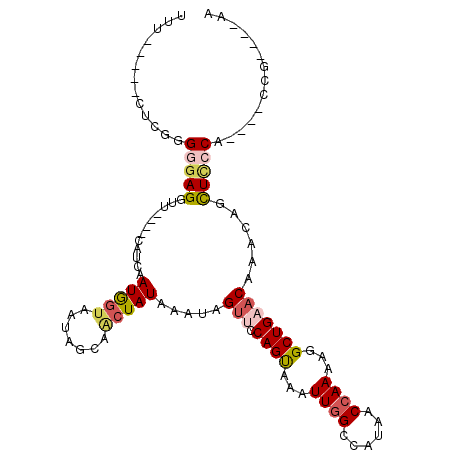
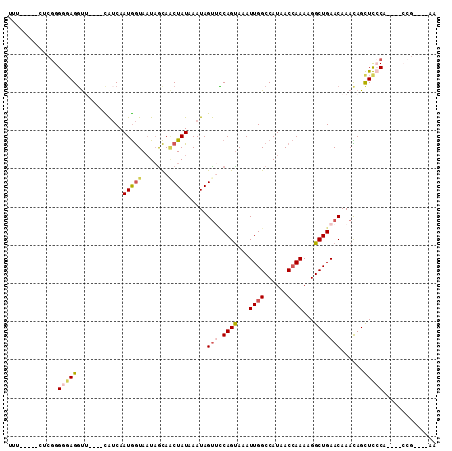
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:28 2006