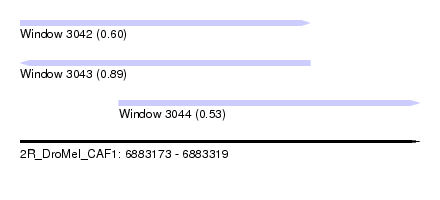
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,883,173 – 6,883,319 |
| Length | 146 |
| Max. P | 0.893178 |

| Location | 6,883,173 – 6,883,279 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.55 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -23.80 |
| Energy contribution | -24.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6883173 106 + 20766785 UAUAUGU----ACCUGAAAGCGGACAAACUCAAACCGCCCGGCCUGAUAUUUUUCGUACGCUGGUACAUAUACAUAUGUAUAUAGAAACUGUACGUUCAUAUGUGUGU--GU (((((((----(((.(...(((((.((..(((..((....))..)))..)).)))))...).))))))))))((((..(((((.(((........))))))))..)))--). ( -28.70) >DroSim_CAF1 2829 108 + 1 UAUAUGU----ACCUGAAAGCGGCCAAACUCAAACCGCCCGGCCUGAUAUUUUUCGUACGCUGGUACAUAUACAUAUGUAUAUAGAAACUGUACGUACAUAUGUGUGUGUGU .......----........((((...........))))(((((.(((......)))...)))))(((((((((((((((((((((...))))..))))))))))))))))). ( -32.00) >DroEre_CAF1 4058 110 + 1 UAUAUGUACAUACCUGAAAGCGGCCAAACUCAAACCGCCCGGCCUGAUAUUUUUCGUACGCUGGUACAUAUACAUAUGUAUAUAGAAACUGUACGUACACAUGUGUGU--GU ...................((((...........))))(((((.(((......)))...))))).((((((((((.(((((((((...))))..))))).))))))))--)) ( -26.30) >consensus UAUAUGU____ACCUGAAAGCGGCCAAACUCAAACCGCCCGGCCUGAUAUUUUUCGUACGCUGGUACAUAUACAUAUGUAUAUAGAAACUGUACGUACAUAUGUGUGU__GU ...................((((...........))))(((((.(((......)))...)))))...((((((((((((((((((...))))..)))))))))))))).... (-23.80 = -24.47 + 0.67)

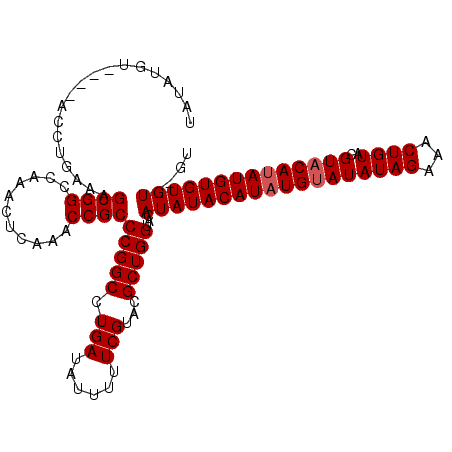
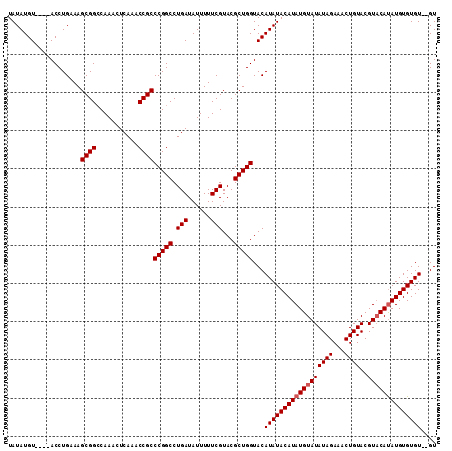
| Location | 6,883,173 – 6,883,279 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 94.55 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -29.09 |
| Energy contribution | -29.20 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.04 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6883173 106 - 20766785 AC--ACACACAUAUGAACGUACAGUUUCUAUAUACAUAUGUAUAUGUACCAGCGUACGAAAAAUAUCAGGCCGGGCGGUUUGAGUUUGUCCGCUUUCAGGU----ACAUAUA ..--....((((((((((.....)))........)))))))(((((((((((((.(((((.....(((((((....))))))).))))).))))....)))----)))))). ( -33.00) >DroSim_CAF1 2829 108 - 1 ACACACACACAUAUGUACGUACAGUUUCUAUAUACAUAUGUAUAUGUACCAGCGUACGAAAAAUAUCAGGCCGGGCGGUUUGAGUUUGGCCGCUUUCAGGU----ACAUAUA ........(((((((((..((.......))..)))))))))(((((((((((((..((((.....(((((((....))))))).))))..))))....)))----)))))). ( -37.20) >DroEre_CAF1 4058 110 - 1 AC--ACACACAUGUGUACGUACAGUUUCUAUAUACAUAUGUAUAUGUACCAGCGUACGAAAAAUAUCAGGCCGGGCGGUUUGAGUUUGGCCGCUUUCAGGUAUGUACAUAUA ..--......((((((((((((......(((((((....)))))))....((((..((((.....(((((((....))))))).))))..)))).....)))))))))))). ( -36.40) >consensus AC__ACACACAUAUGUACGUACAGUUUCUAUAUACAUAUGUAUAUGUACCAGCGUACGAAAAAUAUCAGGCCGGGCGGUUUGAGUUUGGCCGCUUUCAGGU____ACAUAUA ........(((((((((..((.......))..)))))))))(((((((((((((..((((.....(((((((....))))))).))))..))))....)))....)))))). (-29.09 = -29.20 + 0.11)

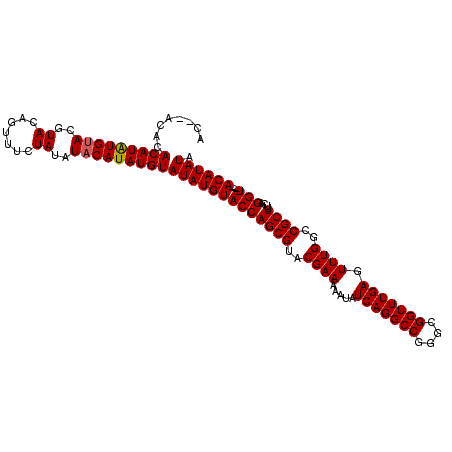

| Location | 6,883,209 – 6,883,319 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 97.01 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -27.92 |
| Energy contribution | -28.37 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6883209 110 + 20766785 GGCCUGAUAUUUUUCGUACGCUGGUACAUAUACAUAUGUAUAUAGAAACUGUACGUUCAUAUGUGUGU--GUGUAUGUCAGUUGGAGUGCGGUCGGCAAAUUGCAGCGAGAA ((((.((......))((((.((..((((((((((((..(((((.(((........))))))))..)))--))))))))..)..)).)))))))).((.....))........ ( -33.00) >DroSim_CAF1 2865 112 + 1 GGCCUGAUAUUUUUCGUACGCUGGUACAUAUACAUAUGUAUAUAGAAACUGUACGUACAUAUGUGUGUGUGUGUGUGUCAGUUGGAGUGCGGUCGGCAAAUUGCAGCGAGAA ((((.((......))((((((((..(((((((((((..(((((....((.....))..)))))..)))))))))))..))))....)))))))).((.....))........ ( -35.50) >DroEre_CAF1 4098 110 + 1 GGCCUGAUAUUUUUCGUACGCUGGUACAUAUACAUAUGUAUAUAGAAACUGUACGUACACAUGUGUGU--GUGUGUGUCAGUUGGAGUGCGGUCGGCAAAUUGCAGCGAGAA ((((.((......))((((((((..(((((((((((((((((.......)))))(....)..))))))--))))))..))))....)))))))).((.....))........ ( -32.80) >consensus GGCCUGAUAUUUUUCGUACGCUGGUACAUAUACAUAUGUAUAUAGAAACUGUACGUACAUAUGUGUGU__GUGUGUGUCAGUUGGAGUGCGGUCGGCAAAUUGCAGCGAGAA .(((((((((.....((((....))))((((((((((((((((((...))))..))))))))))))))......)))))))......((((((......))))))))..... (-27.92 = -28.37 + 0.45)

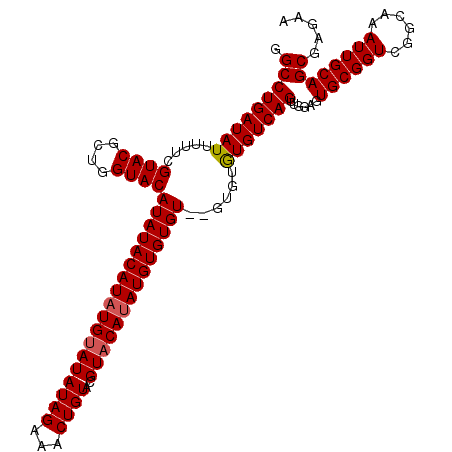
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:17 2006